I'm thrilled to announce @teresa_omeara and I have a new preprint: DeORFanizing Candida albicans Genes using Co-Expression https://www.biorxiv.org/content/10.1101/2020.12.04.412718v1
@teresa_omeara and I have collaborated on 5 papers since grad school, but it was really fun to work on this as our first two-author paper.
We asked if there was enough RNAseq data to build a useful co-expression network for Candida albicans for gene function prediction. It turned out to work amazingly well!
We're working on getting the Candida Albicans Co-Expression Network (CalCEN) out to the community perhaps through FungiDB. Meanwhile, send us your favorite gene and we'll analyze it for you :D
Several years ago I made a network between proteins based on their ligand similarity. @JesseAGillis and @SaraBallouz taught us how to do guilt-by-association gene function prediction and many of the pitfalls to watch out for https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0160098
For example, there are few multi-functional genes like p53, HSP90, etc that tend to be involved in lots of functions. If a network simply predicts these for all functions it does pretty well for retrospective gene function prediction. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0017258
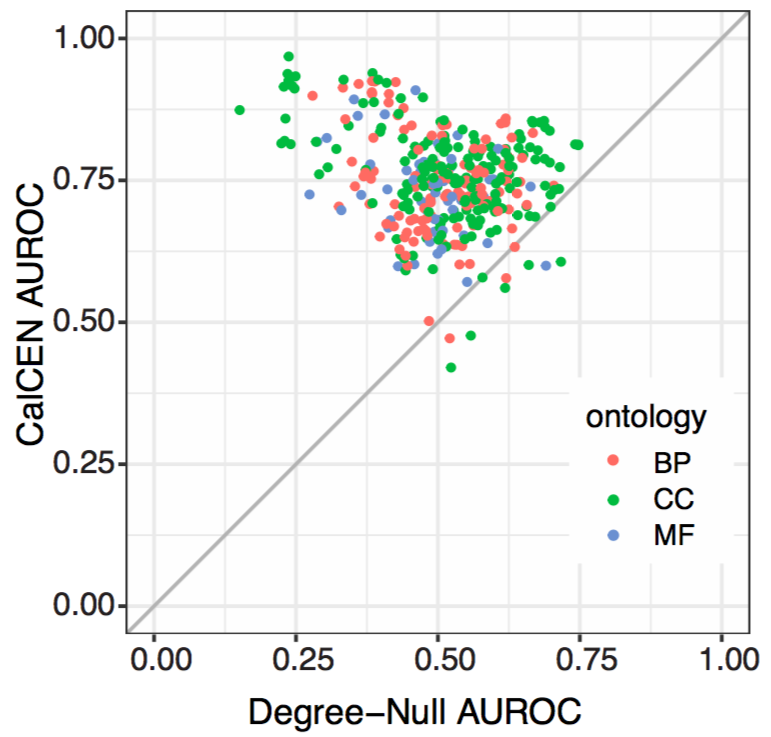
The trouble is, just predicting multi-functional genes isn't very useful for finding new genes for a given function or new functions for a given gene! To test for this bias in a network, simply predicting genes by their network degree for all functions--the Degree Null Predictor.
While in @teresa_omeara was in the @CowenLab she had several really cool functional genomics projects for Candida albicans including screening deletion collection screens and building protein-protein interactions, and I helped with the bioinformatic analysis.
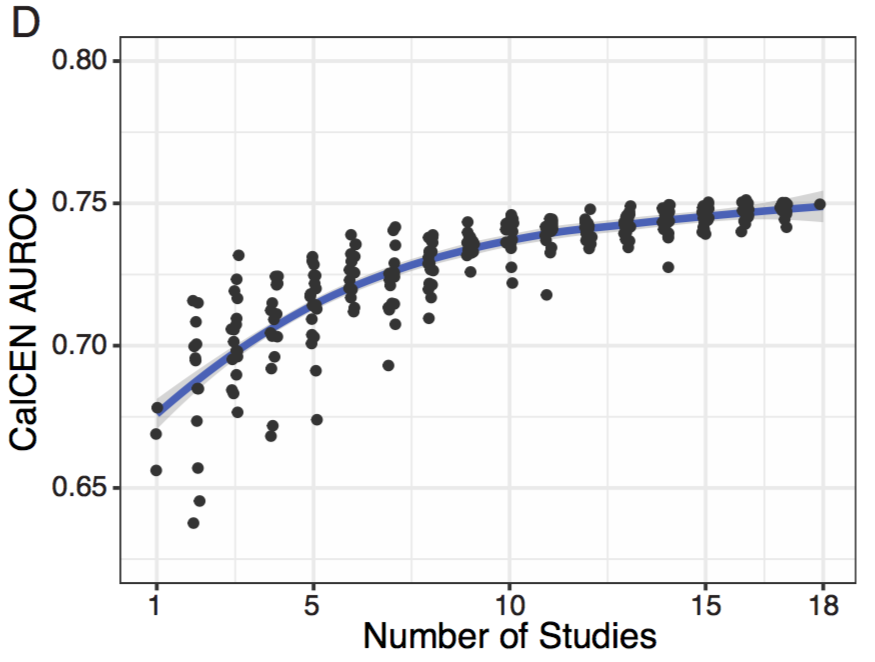
Would Co-expression would complement these studies? There are 18 large scale RNAseq studies in Candida albicans, is this enough to make a useful co-expression network? It looks like ~10 are needed, and the performance hasn't saturated by 18 is plenty but more would help.
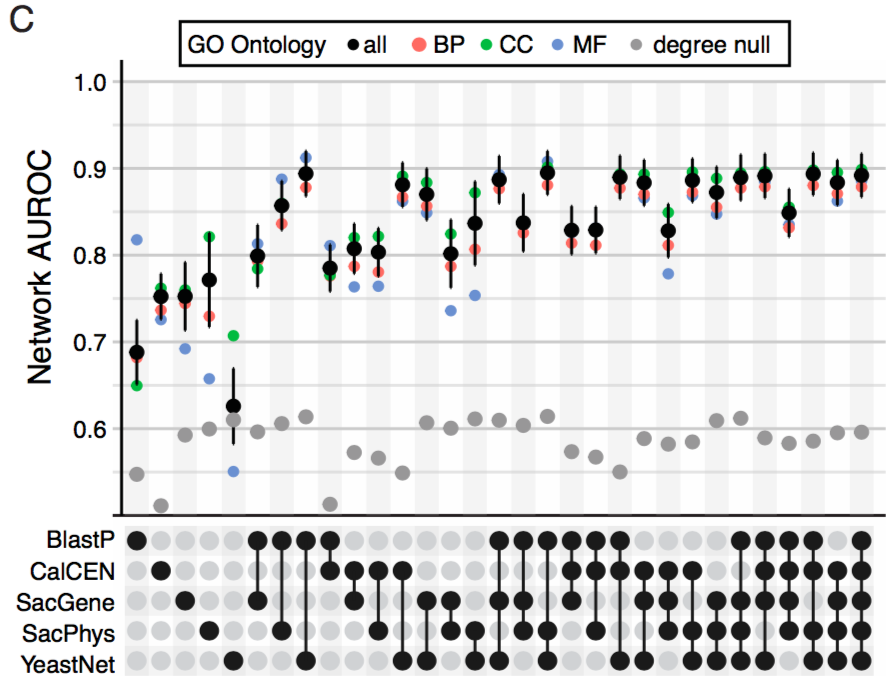
Comparing Co-expression to other networks, we see that CalCEN has strong predictive accuracy with very low multifunctionality bias. Further when combined with other networks it adds more signal.
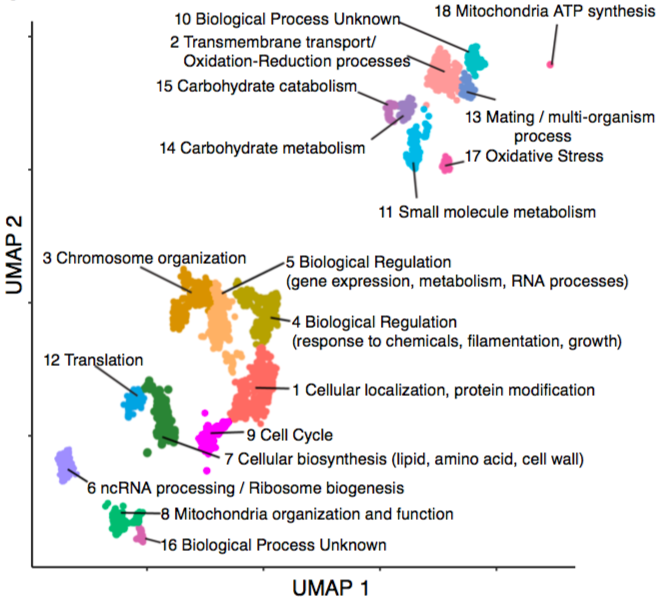
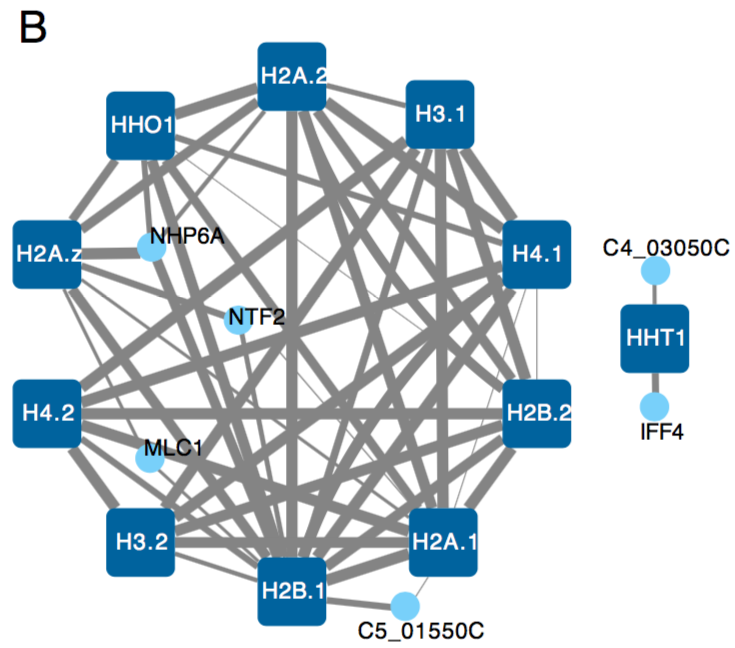
To explore the network, we looked first at known gene clusters. Eg. Histones proteins cluster except for HHT1, consistent with recent findings in https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000422
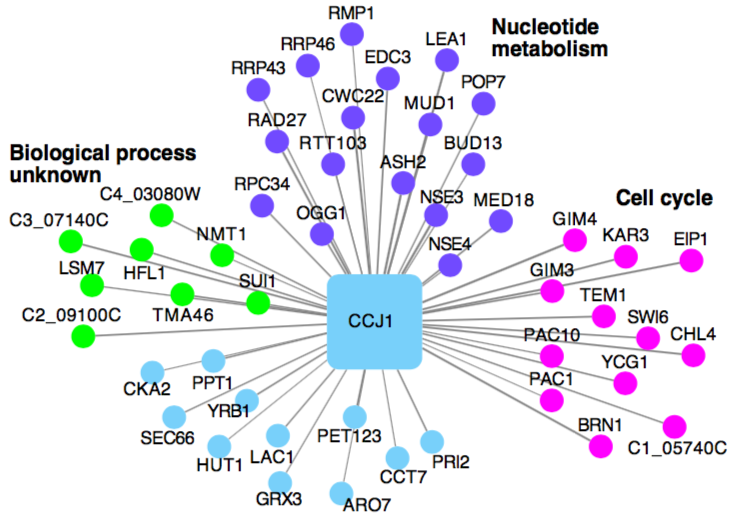
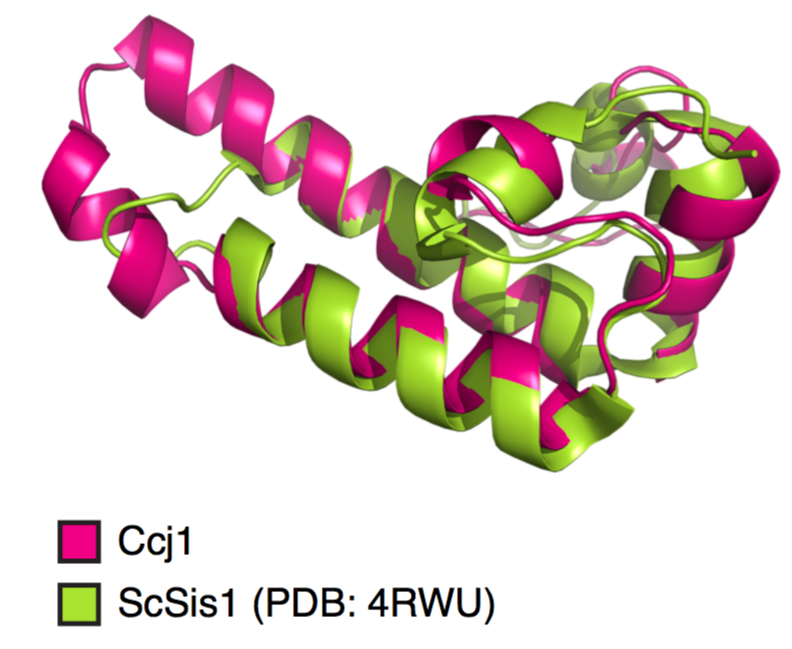
Using deep learning de novo structure prediction with TrRosetta, we verify that it has a DnaJ domain that is similar to the solved structure for SIS1 in Sac.

 Read on Twitter
Read on Twitter