Thrilled to share our pre-print describing single-cell, TCR/BCR repertoire analyses, serum proteomics, and functional studies in MIS-C where we define features that help elucidate mechanisms of immune-mediated tissue damage. Here's a summary. (1/n)
https://www.medrxiv.org/content/10.1101/2020.12.01.20241364v1
https://www.medrxiv.org/content/10.1101/2020.12.01.20241364v1
What is MIS-C? A rare, ‘multisystem inflammatory syndrome in children’ occurring weeks after mild/asymptomatic infection with SARS-CoV-2. These kids develop fever, abdominal pain, rash, and often shock. MIS-C overlaps with Kawasaki (e.g., work by Mike Levin, @BrodinPetter). (2/n)
We assessed 15 MIS-C patients and clinically defined severe (MIS-C-S) vs moderate (-M) based on the requirement for vasoactive medication and/or positive pressure ventilatory support. This stratification turned out to be key for new insights I'll describe below. (3/n)
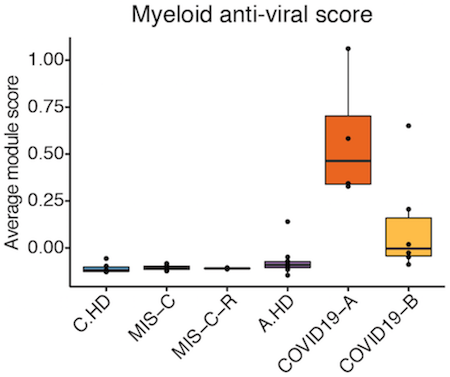
Finding #1: MIS-C patients are + for anti-SARS-CoV-2 antibodies but not necessarily viral RNA. We also found that the anti-viral gene signature in #COVID19 patients is not seen in MIS-C. Still unclear if MIS-C could be set off by another post-viral microbial trigger (gut?). (4/n)
Finding #2: Serum proteomics in MIS-C vs child healthy donor (C.HD) highlights cytokine storm, fluid shear stress, and coagulation pathways. How does innate immunity contribute? (5/n)
…Two potential clues: elevated myeloid S100A alarmins (inflammation amplifiers) and elevated NK/CD8 cytotoxicity genes. MIS-C-R = post-recovery. (6/n)
Finding #3: Are there autoantibodies? Support for this possibility first came from serum antibody screens for reactivity to human proteins (see @BrodinPetter, @BogunovicLab work). We found elevated plasmablasts in MIS-C, and increased IgG1+ cells. (7/n)
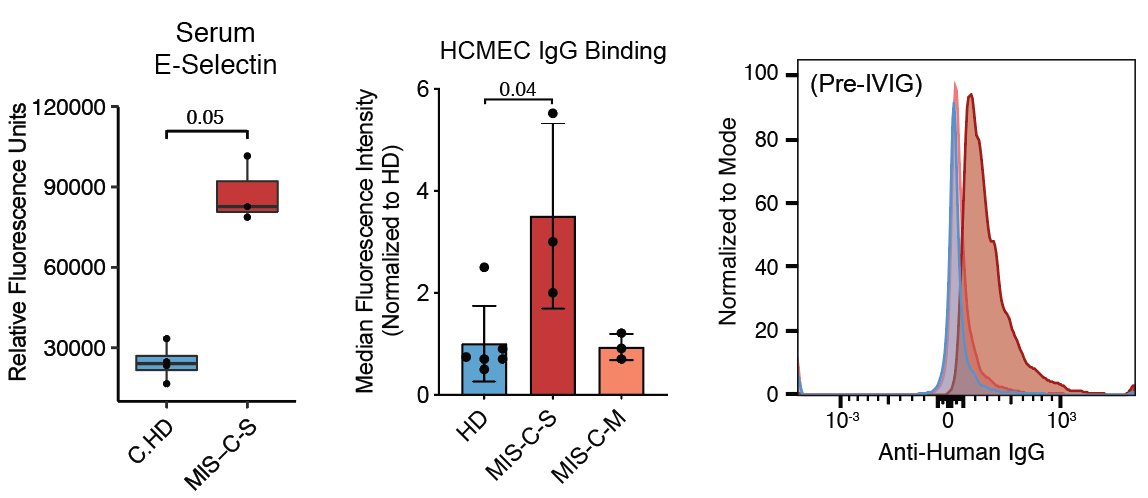
…Next is where severe vs moderate comes in. We see more pronounced plasmablast phenotypes and elevated serum E-selectin (endothelial) in MIS-C-S. Importantly, we also detect binding of serum IgG from severe patients to cultured cardiac endothelial cells. (8/n)
What about TCR reactivity? We don’t know yet. Intriguingly, we found a significant increase in Vb11-2 TCRs in severe MIS-C, also in the recent preprint below. So far, we don’t know if TCR skewing occurs before (from SARS-CoV-2?) or after MIS-C onset. (9/n) https://www.biorxiv.org/content/10.1101/2020.11.09.372169v1.full-text
Lots more to do. We are working hard to increase numbers of analyzed patients and are mapping out potential mouse models for further mechanistic studies. What’s the EC antigen (protein/sugar/other)? Are there genetic variants relevant for MIS-C? Predictive biosignatures? (10/n)
Many thanks to an incredible team @YaleMed @YaleIBIO @YNHH! Special shout out to two talented scientists in my lab, Anjali Ramaswamy (PhD student) and Nina Brodsky (pediatric intensivist/postdoc), with support from Andrew Rice (lab manager). (11/n)
Generous collaborators include: David Hafler lab (esp. Tomo Sumida), Ric Pierce, @david_van_dijk, @skleinstein, Xiting Yan, John Tsang, @KaminskiMed, @RamiUnterman, and others. Helpful EC advice from Jordan Pober. (12/12)

 Read on Twitter
Read on Twitter