So the next new Covid-19 testing technology to look at is the OPTIGENE LAMP test - as covered here in the Guardian.
1/13
Experts question claimed accuracy of Covid-19 saliva tests https://www.theguardian.com/world/2020/dec/04/experts-question-claimed-accuracy-of-covid-19-saliva-tests?CMP=share_btn_tw
1/13
Experts question claimed accuracy of Covid-19 saliva tests https://www.theguardian.com/world/2020/dec/04/experts-question-claimed-accuracy-of-covid-19-saliva-tests?CMP=share_btn_tw
The main body of the report is here, appendicies with data tables can be obtained by email
2/13 https://www.gov.uk/government/publications/rapid-evaluation-of-optigene-rt-lamp-assay-direct-and-rna-formats/rapid-evaluation-of-optigene-rt-lamp-assay-direct-and-rna-formats
2/13 https://www.gov.uk/government/publications/rapid-evaluation-of-optigene-rt-lamp-assay-direct-and-rna-formats/rapid-evaluation-of-optigene-rt-lamp-assay-direct-and-rna-formats
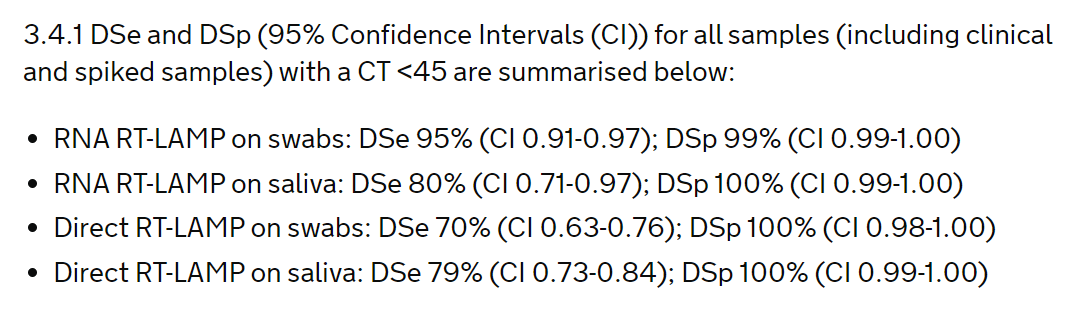
The study looked at using the test on RNA extracted from swabs and saliva, and directly on swabs and saliva. Key results are here:
3/13
3/13
So false negative rates increase from 5% with RNA extracted from swabs (which looks good), to 21% with direct saliva (not so good). But the Government said that missing 21% is OK.
I really wonder what their criteria for good are. Let me know if you've seen then stated.
4/13
I really wonder what their criteria for good are. Let me know if you've seen then stated.
4/13
But there's a twist, linked to that word "spiked". Its not normal to put spiked samples in a clinical study. Not at all normal. Clinical studies are all about knowing how well a test works in people like us, not manufactuered samples
5/13
5/13
59 spiked samples were in the direct saliva group. Despite most of them having low (Ct>=33) viral loads, the test detected the virus in 91% of them. Of the real samples with similar viral loads virus was only detected in 13% of them. So spiked samples are different
6/13
6/13
I corresponded with the authors at the beginning of the week suggesting they presented results without these, as I didn't want to have to write more critical tweets or get quoted in the Guardian saying somebody had done something bad AGAIN.
7/13
7/13
I suggested it would have been much better just to report results from real people. But they haven't done it.
So PLEASE can somebody do a test evaluation study which I can tweet really nice things about? That part of my vocabulary is severely underused at the moment.
8/13
So PLEASE can somebody do a test evaluation study which I can tweet really nice things about? That part of my vocabulary is severely underused at the moment.
8/13
So I've removed the spiked results for you.
Removing them gives an overall sensitivity for direct saliva of 76% - so 24% of cases missed. Compared to 95% - so 5% missed with RNA extracted swabs - so 5 times as many get missed.
9/13
Removing them gives an overall sensitivity for direct saliva of 76% - so 24% of cases missed. Compared to 95% - so 5% missed with RNA extracted swabs - so 5 times as many get missed.
9/13
Now grouped by viral load
Sensitivity for direct saliva is
94% if Ct<25; 71% if 25<=Ct<33; 13% if Ct>=33,
compared to
100% if Ct<25, 95% if 25<=Ct<33; 74% if Ct>=33
for RNA extracted swabs.
10/13
Sensitivity for direct saliva is
94% if Ct<25; 71% if 25<=Ct<33; 13% if Ct>=33,
compared to
100% if Ct<25, 95% if 25<=Ct<33; 74% if Ct>=33
for RNA extracted swabs.
10/13
Get the Appendicies and you will also see worrying variation in sensitivity between the centres for direct saliva
Birmingham 100% (n=16)
Southampton 87% (n=76)
Hampshire 84% (n=19)
Lighthouse lab 54% (n=41)
Manchester 47% (n=15)
11/13
Birmingham 100% (n=16)
Southampton 87% (n=76)
Hampshire 84% (n=19)
Lighthouse lab 54% (n=41)
Manchester 47% (n=15)
11/13
These data suggest a large (5 fold) loss of sensitivity using LAMP on direct saliva, and worrying substantial unexplained differences between centres.
And the test really can't detect when viral levels are low and misses over a quarter when viral levels are moderate
12/13
And the test really can't detect when viral levels are low and misses over a quarter when viral levels are moderate
12/13
But I am still pondering how the Technical Validation Group thinks its OK to throw 59 spiked samples into a "clinical study". The DHSC also says there's nothing wrong in doing that.
That certainly wouldn't get past peer review at any reputable journal I deal with.
13/13
That certainly wouldn't get past peer review at any reputable journal I deal with.
13/13

 Read on Twitter
Read on Twitter