Did you tune the MPI ranks of your MD simulations for the best simulation performance? Using MDBenchmark we show how the optimal number of MPI ranks changes with the number of atoms in 23 exemplary systems using GROMACS @GMX_TWEET @AIP_Publishing
https://aip.scitation.org/doi/figure/10.1063/5.0019045
1/10
https://aip.scitation.org/doi/figure/10.1063/5.0019045
1/10
Biomolecular researchers love to simulate water  ! While most of the systems are composed of solvent, some have significant amounts of membranes, dense protein solutions or other solutes such as DNA/RNA.
! While most of the systems are composed of solvent, some have significant amounts of membranes, dense protein solutions or other solutes such as DNA/RNA.
2/10
 ! While most of the systems are composed of solvent, some have significant amounts of membranes, dense protein solutions or other solutes such as DNA/RNA.
! While most of the systems are composed of solvent, some have significant amounts of membranes, dense protein solutions or other solutes such as DNA/RNA.2/10
We show that it is best to use as many MPI ranks as available physical/logical cores on your node when simulations are run on CPU-only nodes. Actually, you should also enable hyperthreading to get that sweet 10% performance boost 

3/10


3/10
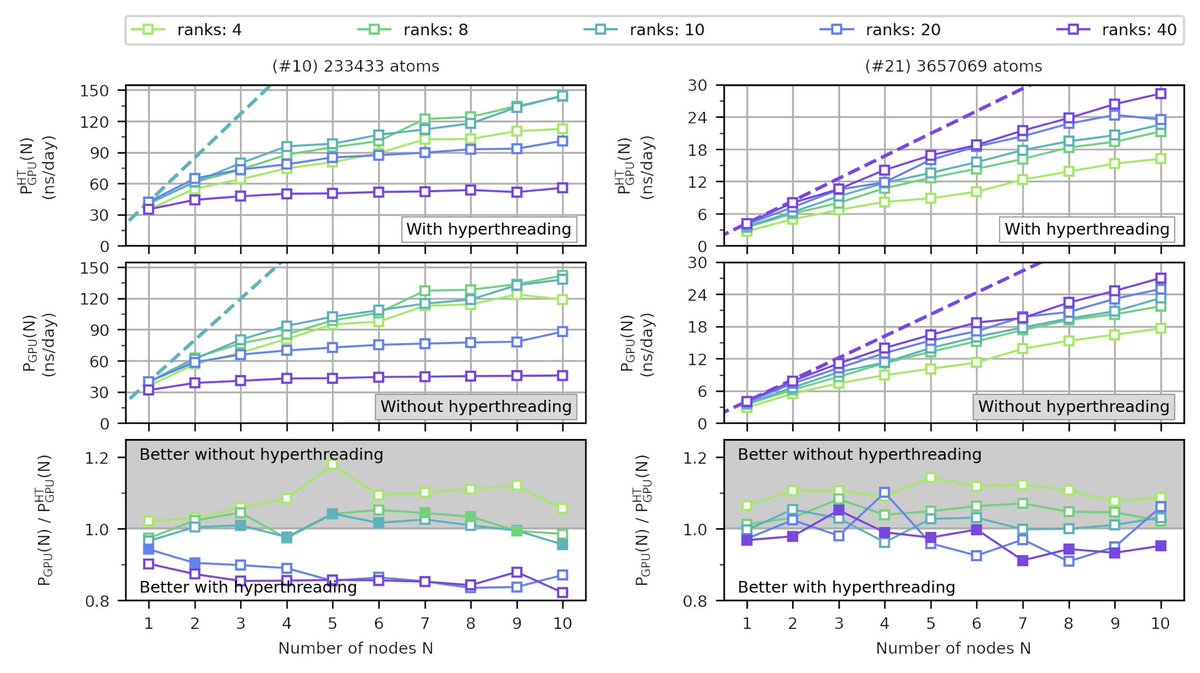
In contrast, on mixed CPU-GPU nodes the number of ranks depends on the system size. Smaller systems achieve their best performance with few MPI ranks, whereas bigger systems require more ranks Hyperthreading is not necessarily beneficial with GPUs.
Hyperthreading is not necessarily beneficial with GPUs.
4/10
 Hyperthreading is not necessarily beneficial with GPUs.
Hyperthreading is not necessarily beneficial with GPUs.4/10
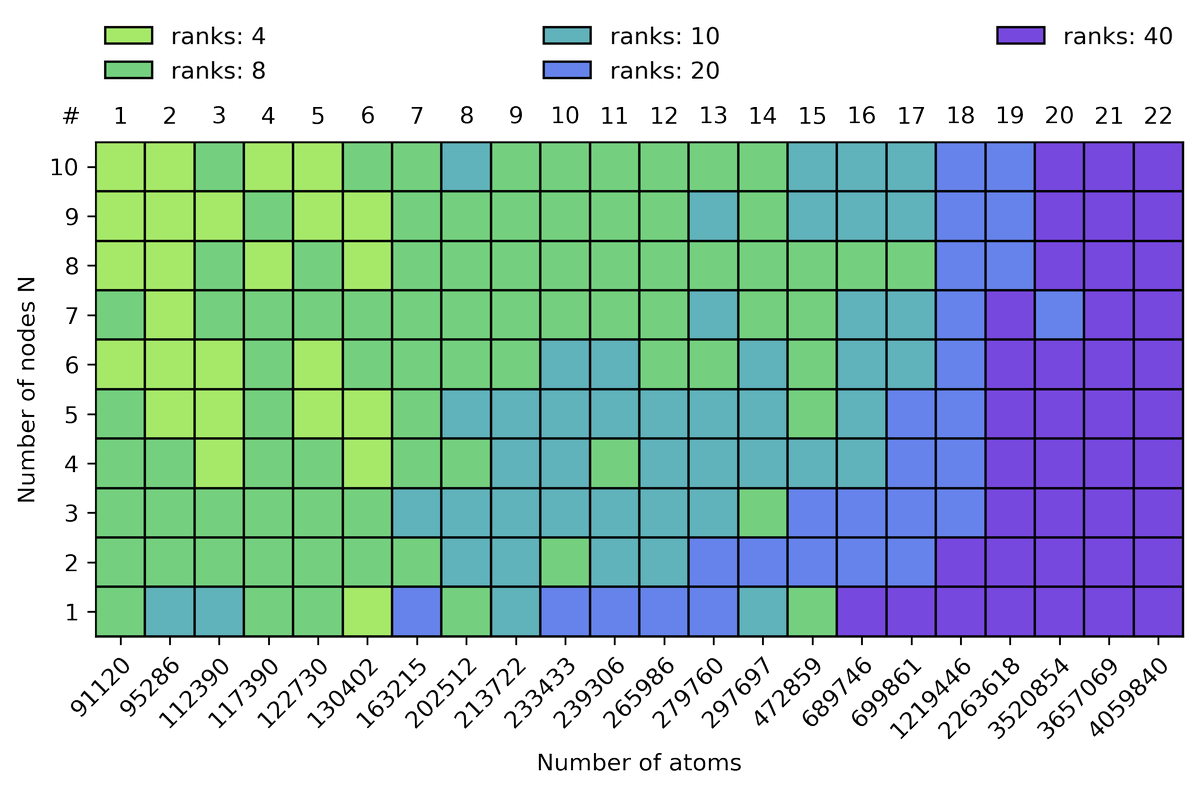
Repeating this exercise for all our tested systems, we can summarize the observations as follows: (i) the number of MPI ranks decrease with increasing node numbers and (ii) the number of MPI ranks increase with increasing system size.
5/10
5/10
Are mixed CPU-GPU nodes always the best choice? No! If you have sufficiently big systems, you can achieve higher absolute performances on CPU-only nodes. These are not accessible on mixed CPU-GPU nodes. Also we show how Amdahl's law helps us to to model our observations.
6/10
6/10
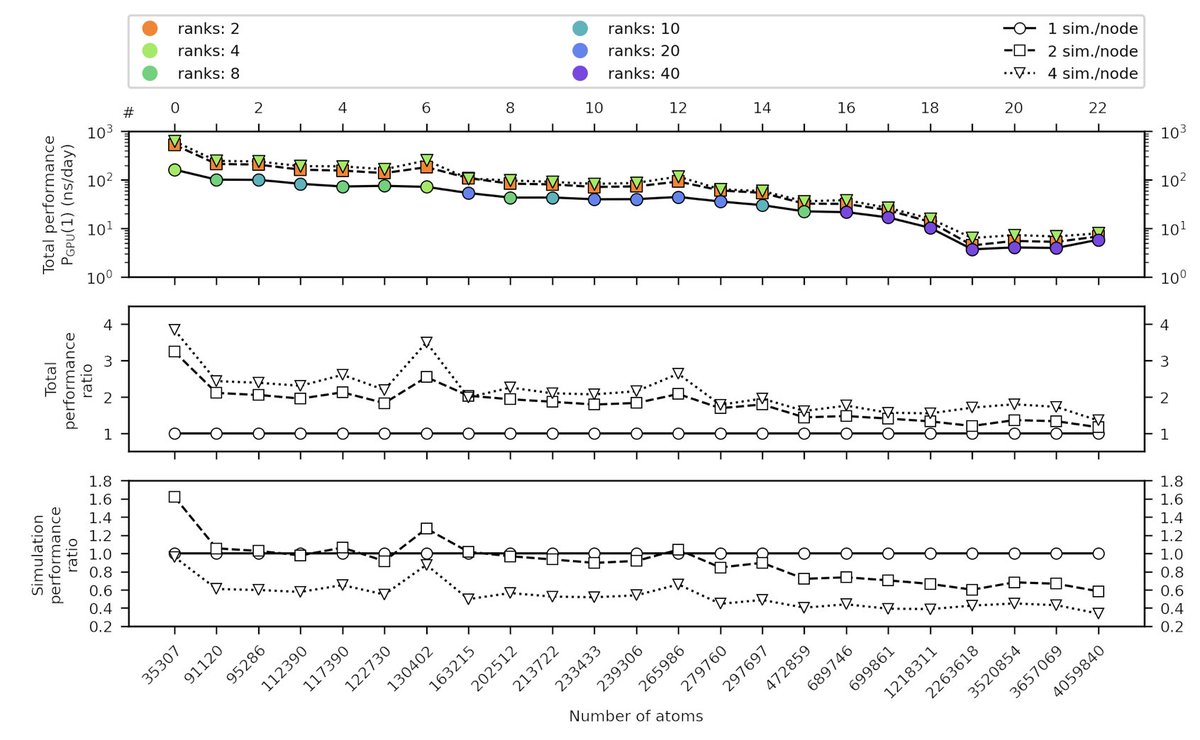
But wait there is more... computational resources than you thought! Did you know that you can run multiple simulations on a single node? On mixed CPU-GPU nodes this will actually increase your total simulation performance by 1.5- to 4-fold!
7/10
7/10
How did we produce all this data? We used our awesome MDBenchmark Python package! Version 3 is available on PyPI and conda-forge. It allows you to scale MD systems across different node numbers and try out different numbers of MPI ranks.
8/10
8/10
You can find the code on GitHub: https://github.com/bio-phys/mdbenchmark. We are all ears for any issues you encounter, as well as contributions of new features!
The documentation is hosted over at @readthedocs: https://mdbenchmark.readthedocs.io/en/latest/
9/10
The documentation is hosted over at @readthedocs: https://mdbenchmark.readthedocs.io/en/latest/
9/10
It was a pleasure to work on this project with @MSiggel, @MaxLinke4, Gerhard Hummer and @jkoefinger! Big shout out the MPCDF in Garching: Klaus Reuter, Markus Rampp and Sebastian Kehl. And also a big thank you to all members of TB @MPIbp for providing their MD systems 
10/10

10/10
11/10

 Read on Twitter
Read on Twitter