Excited to share our new preprint! We integrated 100+ single-cell/nucleus RNAseq datasets (23 new!) to explore rare cell states in myogenesis. ~350k cells/nuclei and the first spatial RNAseq data in regenerating muscle! https://www.biorxiv.org/content/10.1101/2020.12.01.407460v1
Muscle stem cells are rare (~1% of cells)- we figured that rather than physically isolating cells (FACS), we could digitally isolate them. To do that, we needed a lot of data though...
We started by generating ~95k new cells, spread across the first week of injury response. We added in more data previously generated by @AndreaDeMicheli ( https://www.cell.com/cell-reports/fulltext/S2211-1247(20)30235-7) but it wasn't enough!
We then realized that we could use publicly available data to supplement what we had! So we downloaded 79 datasets (from fastqs), which span a variety of ages, injury models, injury timepoints, etc. After a LOT of QC, we integrated them with Harmony
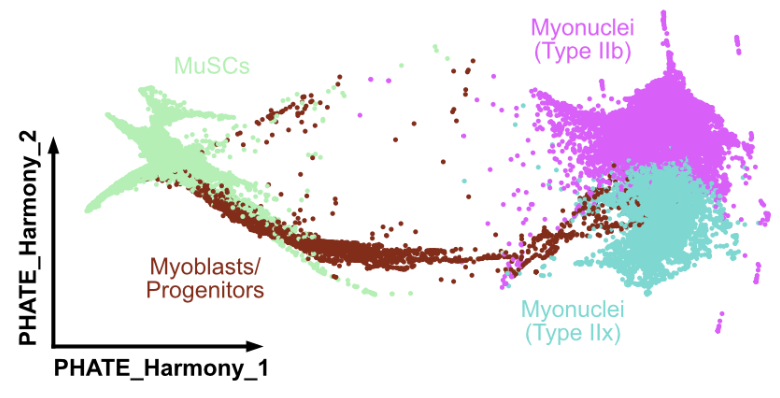
After integration, we subset out the ~80k myogenic cells and then used PHATE to perform pseudotime analysis. The exciting thing here is that we can see every step of MuSC differentiation!!
For the first time, we could now properly see the intermediate cell states (Myog+, Mymx+, Mymk+) in myogenesis! With some simple dif. gene expression, we found transcr. factors and surface markers that define myogenic commitment
The big thing I want to mention here is that, at the RNA level, standard MuSC markers (esp. Itga7, Cxcr4, and Itgb1) are expressed in committed cells! This could explain poor engraftment of isolated MuSCs!
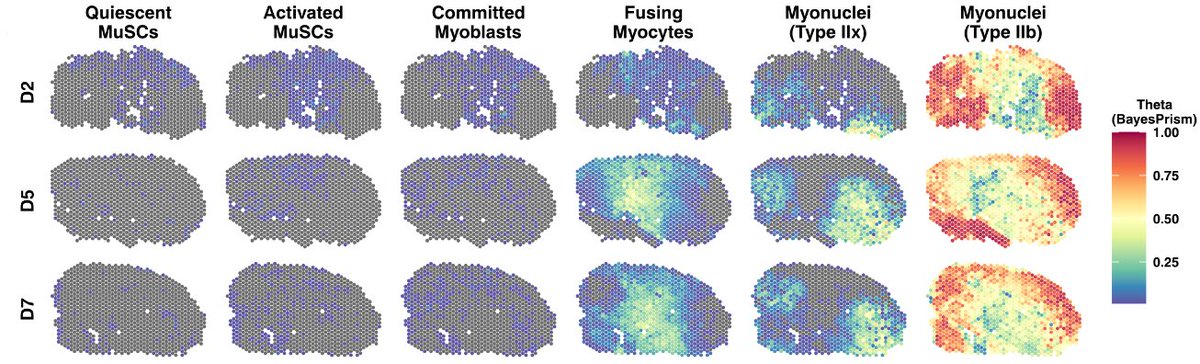
Last, we used the 350k dataset as a reference to deconvolve Visium data, collected at 3 timepoints after injury. Each spot here contains ~5-15 cells, and with BayesPrism (from @tinyichu & @charlesdanko) we can see what those cells are!
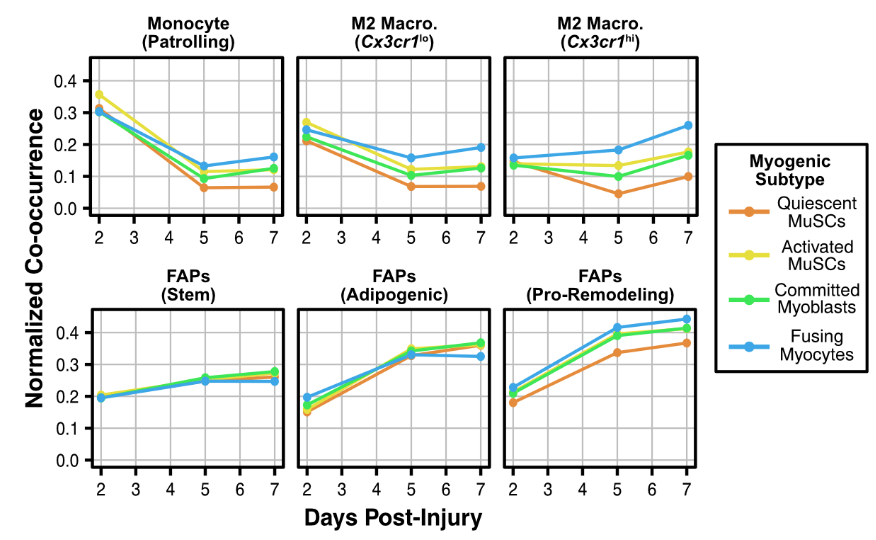
With the deconvolved spots, we simply looked at which cells occur together most often, and plotted the co-occurrence over injury response. Because we used such a deeply covered reference, we can see cell subtype colocalization!
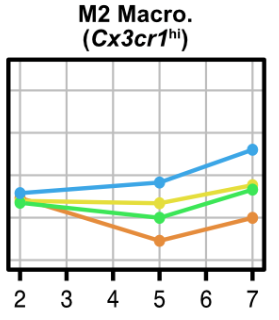
This result got me especially excited- look at how this subset of M2 macro's differentially co-localizes with different myogenic cell states (quiescent vs. activated vs. fusing). This to me says that there is a specific interaction here that triggers maturation
This preprint is really just a first step- we are still working to get a web resource together so that others can explore this dataset. Still lots of cool questions to answer! We are also still adding more data, so message me if you want to include yours in v2!
The high level idea here is that batch-correction tools (harmony, scanorama, BBKNN, etc) now enable these massive scales of analysis. Anyone with a decent amount of RAM can replicate projects like the Tabula Muris on their own by crowd-sourcing data!
Big thanks to all the co-authors, especially @bdcosgrove and @DeVlaminckLab who let me use an exorbitant amount of compute time to stitch this all together.
Even bigger thanks to everyone who made their data publicly available and made this possible! @doug_millay @jacobkimmel @BrackLab @LeGrandLab1 @sartorellliv @AndreaDeMicheli and so many others!

 Read on Twitter
Read on Twitter