Really happy to finally share results from my postdoc work in the Zipfel lab: how plants respond to a variety of different elicitors and what we can learn from it
1/12 https://www.biorxiv.org/content/10.1101/2020.11.30.404566v1
1/12 https://www.biorxiv.org/content/10.1101/2020.11.30.404566v1
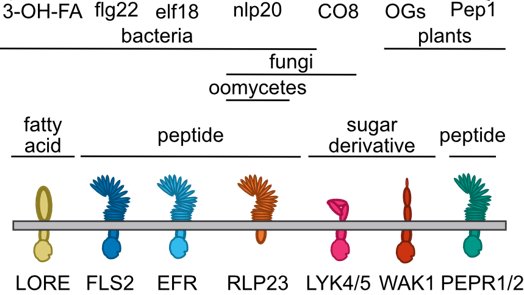
Although we know a lot of molecules plants recognize as damage/pathogen patterns, not many studies have compared more than one of these elicitors, or looked at response at more than one time. We took 7 elicitors, and assayed transcriptome at 6 early time points 0-3 hrs
2/12
2/12
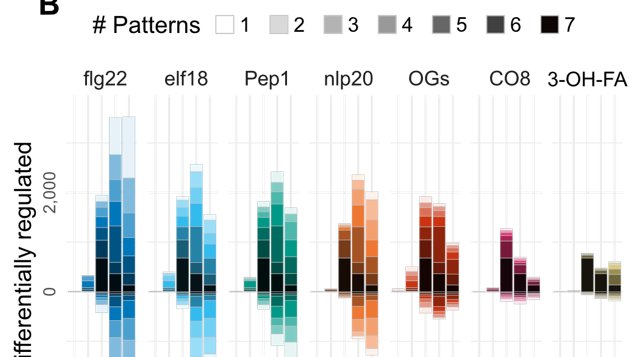
Like we suspected from single elicitor studies, plants change a lot of their transcriptome, and a lot of the changes for each elicitor are shared with many others: almost 1,000 genes induced commonly by all elicitors tested
3/12
3/12
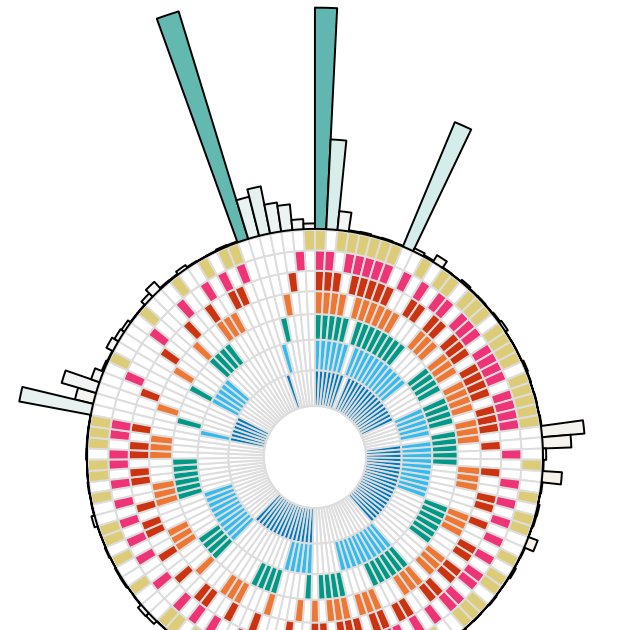
More than that, there aren't strong patterns of common responses among any subgroups of elicitors (like those from plants vs. those from microbes): just the strong trend of all elicitors commonly, or some elicitors uniquely
4/12
4/12
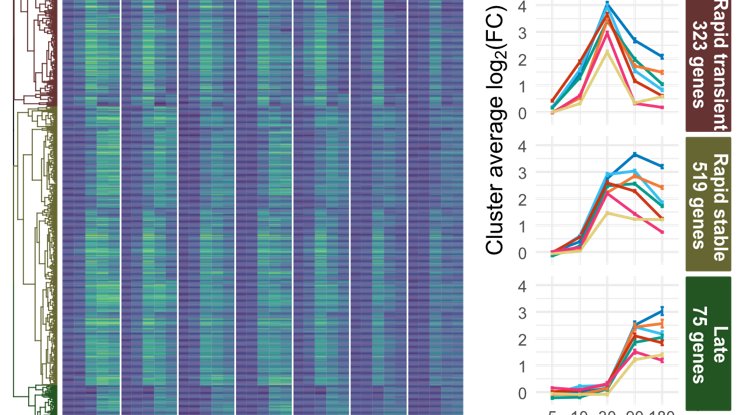
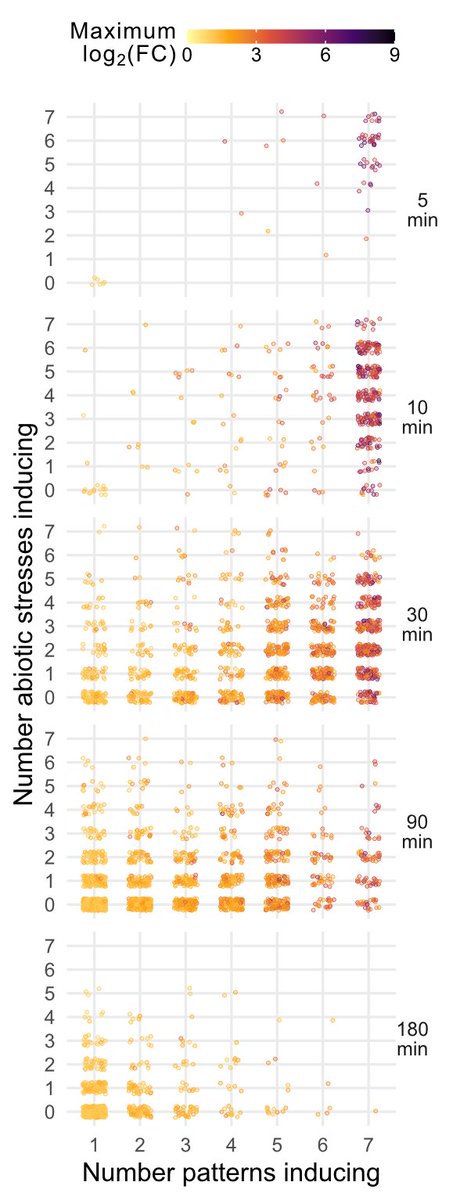
BUT, there are timing patterns, with separation of extremely rapid (5 minutes!) or slow, stable or transient response.
These sets are enriched for different cis elements; while 30 min genes are induced in WRKYs, fast-response genes are enriched for CAMTA TF binding sites.
5/12
These sets are enriched for different cis elements; while 30 min genes are induced in WRKYs, fast-response genes are enriched for CAMTA TF binding sites.
5/12
CAMTAs regulate the plant general stress response; is that what we're seeing? Using the AtGenExpress dataset, we found that rapidly induced genes are induced by many elicitors and many abiotic stresses, with responses becoming more specific over time. (my favorite plot!)
6/12
6/12
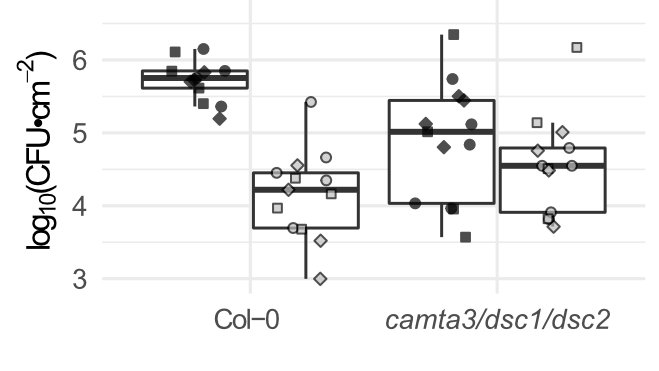
The GSR is major part of elicitor transcriptional response: is it a major part of immunity? Mutants in CAMTA3 (with the two R proteins that guard it) are altered in both basal and elicitor-induced immunity.
7/12
7/12
Responses become more specific over time... are there genes that are induced by all elicitors, and no abiotic stresses? Yes! And, in this time frame, only a few of them! of just 40 core immunity response genes, two are closely related GLR Calcium channels
8/12
8/12
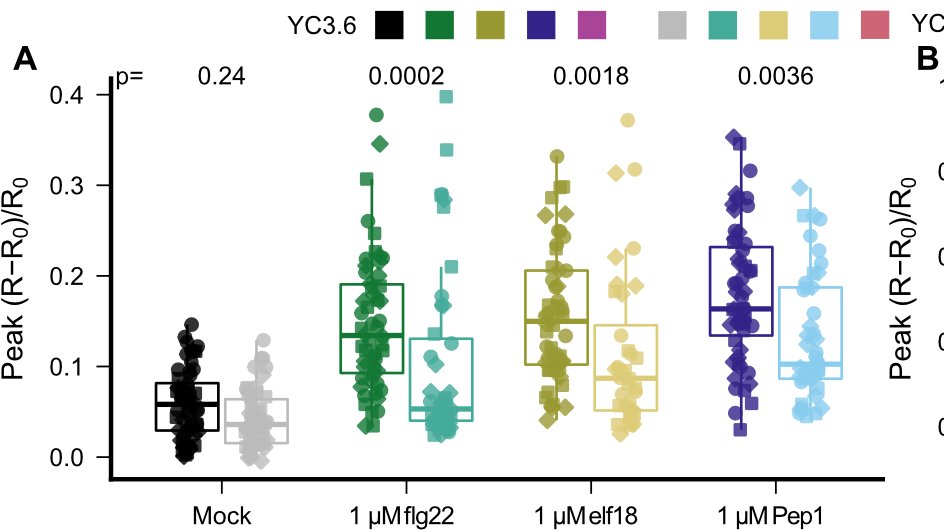
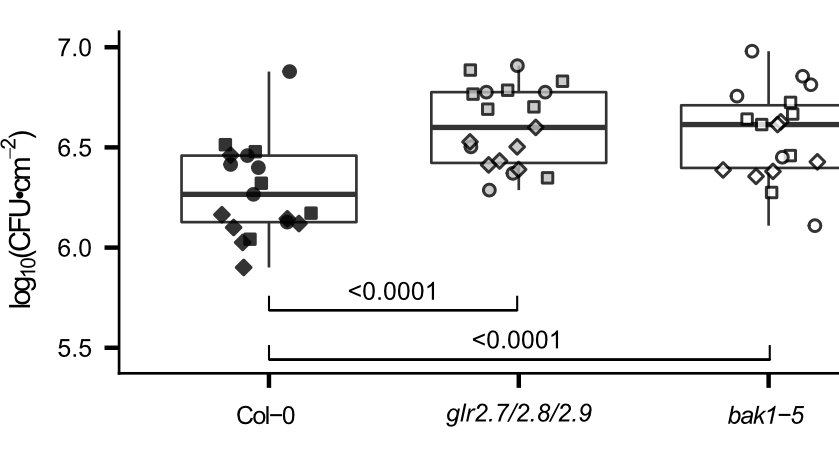
Cutting out GLR2.7, 2.8, and 2.9 by CRISPR makes plants slightly less able to respond to elicitor perception with a Calcium influx... but no change in salt response, in line with elicitor-specific induction.
9/12
9/12
That's not a huge change in Calcium influx, but it is enough to result in quantitative changes in bacteria susceptibility!
10/12
10/12
We're really excited for more possibilities in this dataset - elicitor-specific gene induction, elicitor-specific timing, function of other CIR genes, other transcriptional regulators of elicitor and general responses... and more!
11/12
11/12
Big thanks to many people who made this possible, including @PriyaPimprikar, @ThorstenNuernbe, postdoc communities at TSl and UZH, and many others. It's been a fun trip, hope you enjoyed the quick version and check out BioRxiv if you are still curious!
12/12
12/12

 Read on Twitter
Read on Twitter