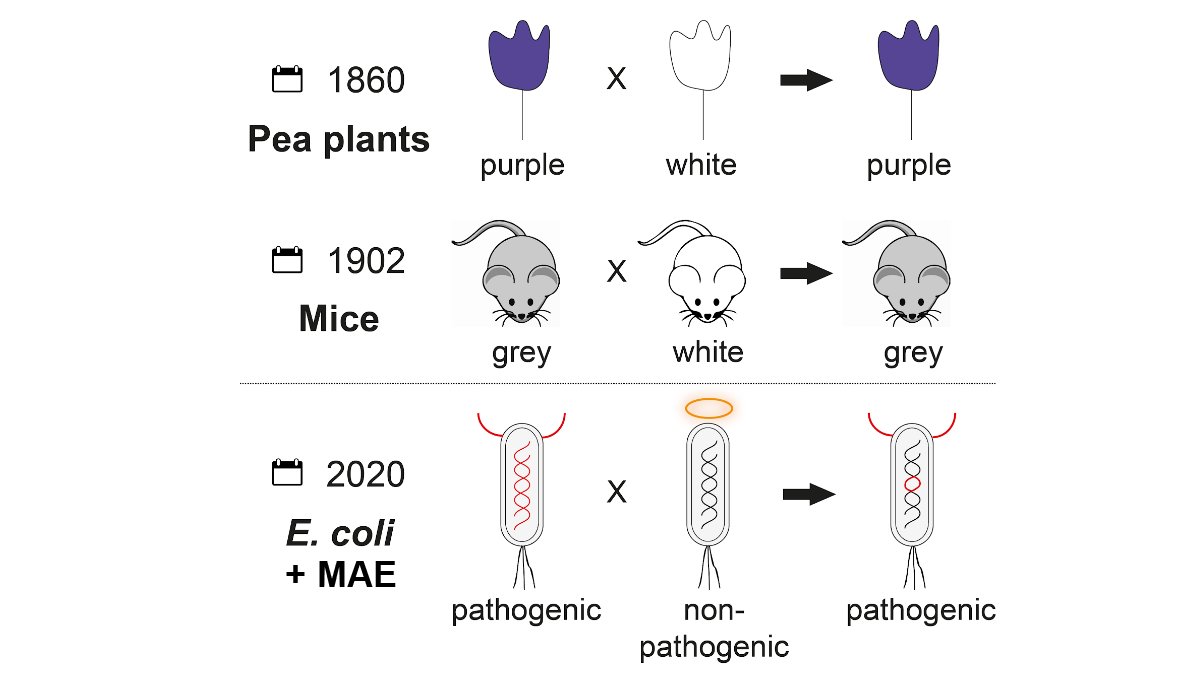
Super excited to share a preprint ( https://www.biorxiv.org/content/10.1101/2020.11.30.405282v1) for one of the lab’s long term, ambitious projects - doing effective, genome-scale, sexual genetics in E. coli. This is making F1 hybrids in bacteria! We call our new technique MAE (Mass Allelic Exchange) - 1/14
You may be thinking, “F1 hybrids in bacteria”?! Bacteria are haploid, but they can do horizontal gene transfer. They also do homologous recombination - the same process that generates random assortment of alleles in pea plants, mice - whatever you associate F1 hybrids with. 2/14
Also, some bacteria are “naturally competent” - they can take up extracellular DNA and incorporate this into their genomes by homologous recombination - Streptococcus pneumoniae and Campylobacter jejuni are some notable examples. But not E. coli! 3/14
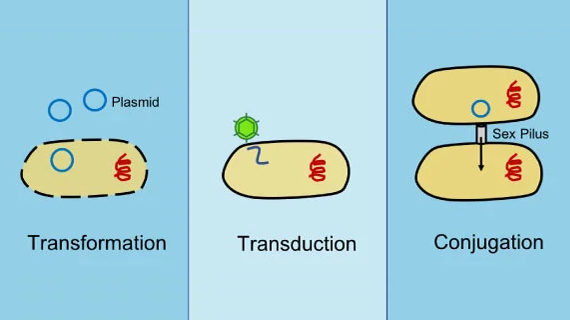
E. coli is not naturally competent, but we have long known that we can transfer alleles between strains, making “sexual hybrids”, using the traditional trinity of transfer techniques: transformation, transduction, or conjugation. 4/14
So why is making F1 hybrids in E. coli a big deal? The traditional techniques typically transfer relatively small parts of the chromosome and often require special selection strategies. It’s hard to make libraries of hybrids that could be used to dissect a complex phenotype. 5/14
A survey of other techniques leads to a wish list which hadn’t yet been fulfilled - until MAE of course!
1. Usable in most E. coli strains, including wild type, clinical isolates
2. Able to transfer large (in the range of 50%) portions of the chromosome
6/14
1. Usable in most E. coli strains, including wild type, clinical isolates
2. Able to transfer large (in the range of 50%) portions of the chromosome
6/14
(wishlist continued)
3. Random recombination breakpoints - just like in peas or mice
4. Efficient enough to generate libraries of thousands of clones (i.e. to get genome-wide coverage)
5. No residual markers or genetic scars
7/14
3. Random recombination breakpoints - just like in peas or mice
4. Efficient enough to generate libraries of thousands of clones (i.e. to get genome-wide coverage)
5. No residual markers or genetic scars
7/14
The key innovation was a strong negative selection system we developed when the lab first started. This provides >1000x stronger negative selection even in cloning strains of E. coli - and doesn’t lose efficiency in clinical strains ( https://pubmed.ncbi.nlm.nih.gov/25800749/ ) 8/14
We incorporated the negative selection cassette with an Hfr transfer origin into a custom transposon. The Hfr origin allows conjugation to transfer large segments of the chromosome - in the megabase range! 9/14
More details in the paper, but the bottom line is we engineered this to generate unmarked hybrids after conjugation. Negative selection + conjugation gives us a big (billion-fold, yes 10^9!!) boost in transfer efficiency for these large chromosomal segments. 10/14
We did several screens, but the big one took advantage of the ability to use clinical strains. We studied how uropathogenic E. coli makes large bacterial collections inside bladder epithelial cells - a key contributor to our inability to cure recurrent UTI with antibiotics. 11/14
Using an in vitro model for bacterial communities, we found that transfer of only one single operon (chu, involved in iron acquisition) was sufficient to enable a cloning strain of E. coli (the venerable MG1655) to form these intracellular collections! 12/14
Please go check out the preprint! We have the first arrayed library of MG1655/UTI89 hybrids, which collectively have nearly 90% of the UTI89 chromosome transferred, and are happy to share this library for other cool screens! 13/14
Link: https://www.biorxiv.org/content/10.1101/2020.11.30.405282v1
Link: https://www.biorxiv.org/content/10.1101/2020.11.30.405282v1
Big congratulations to @VarnicaK who made this dream for doing sexual genetics in E. coli a reality. She developed the negative selection system and solved a litany of issues - finally achieving that billion-fold increase in efficiency! Lots of help also from @liyanaaoy! 14/14

 Read on Twitter
Read on Twitter