Ready for a new "ML pitfalls in genomics w/ Jacob"? When evaluating ML models across cell types/individuals, you MUST baseline against the avg activity or risk being fooled by seemingly good performance. Thrilled to finally see this quick read out! 1/ https://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-02177-y
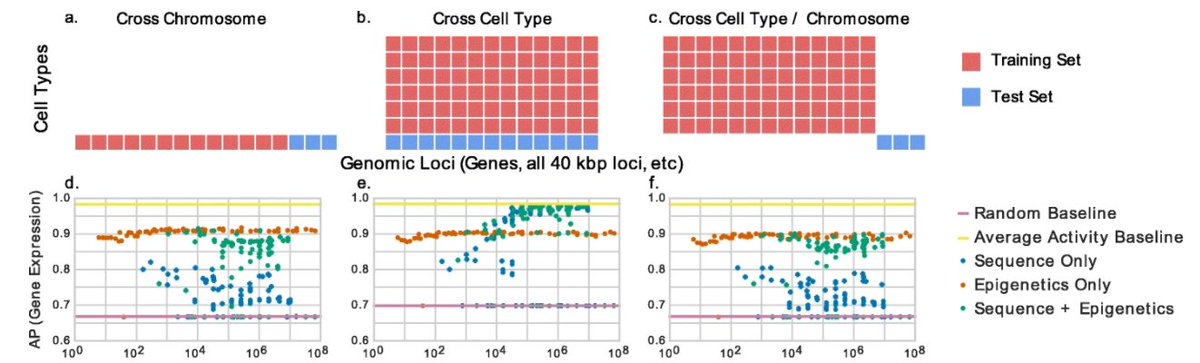
The most common evaluation scheme for genomics data involves training/evaluating on separate chromosomes. However, this is unrealistic because you rarely only have data for some chroms but not others. Others, including @akundaje and @michaelhoffman have also pointed this out. 2/
A more realistic evaluation setting is "cross celltype", where you train a model genome-wide in some biosamples and make predictions genome-wide in other biosamples. This matches reality, where you've either performed a genome-wide experiment in a biosample or you haven't. 3/
As an aside, there is a third setting, which we call the "hybrid" setting, which is both cross-celltype and cross-chromosome. This setting can be useful when generalization to new sequence is important, such as for a model that can generalize across species. 4/
To demonstrate the practical differences in these settings, we trained CNNs/DNNs of increasing complexity to predict gene expression. We observed very different results in the cross cell type setting!
In fact, it looked like models got better as they got increasingly big (up to 10^6 parameters!), AS LONG as they used nucleotide sequence as an input. 6/
That didn't seem biologically plausible to us. Is it really the case that there are super complicated sequence motifs that need 10^6 parameters to model? We called bullshit! ( @CT_Bergstrom gave us permission) 7/
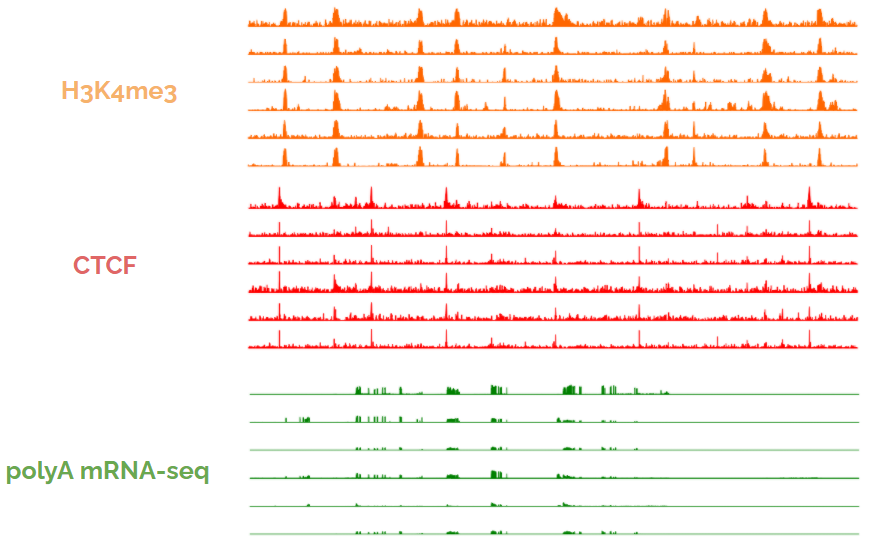
Long story short: because biochemical activity is frequently similar across cell types, using the activity from one cell type to predict activity in another cell type frequently gives good performance. Look at these tracks below, each coming from a different cell type. 8/
We call averaging the activity across the training celltypes the "average activity baseline," and found that super complex ML models were simply using nucleotide sequence as a hash to memorize the average activity. 9/
You'll notice that these models never *outperform* the average activity, they simply *asymptote* to it. 10/
The key takeaway: make sure that you are outperforming the average activity baseline. A convenient property of this is that, by definition, outperforming it means you're making accurate cell type specific predictions. 11/
We felt it was important to write this piece not only to point out this pitfall, but also to encourage authors to carefully consider the predictive setting they are applying their models in. Frequently, papers will imprecisely describe what they did, but it's IMPORTANT. 12/
As you can see here: one can publish a model w/ good performance one year, and even better performance the next year, by replacing one component with a more complex one. Be skeptical of these papers-- they might simply be better memorizing the avg activity. 13/
Read the paper! It's short and describes the issue in more detail than I can here. You can also watch me give a talk about it here: 14/14
p.s. I think it's completely appropriate that the editors decided to use "AAA" as the header name, but I wish they added an H at the end of that to appropriately describe the reaction of people who realize they have fallen into this pitfall.

 Read on Twitter
Read on Twitter