I would like to introduce the beta of a bioimage analysis software called *Mastodon*, that we have been working on with @pietzscht for several years now.
Mastodon is our best effort to address interactive cell tracking in large images.
https://github.com/mastodon-sc/mastodon
thread
1/
Mastodon is our best effort to address interactive cell tracking in large images.
https://github.com/mastodon-sc/mastodon
thread

1/
2/
Mastodon aims at offering:
- automatic tracking,
- semi-automatic tracking,
- and manual editing of tracks
in very large images. They key challenge it addresses is the *size of data*:
Mastodon aims at offering:
- automatic tracking,
- semi-automatic tracking,
- and manual editing of tracks
in very large images. They key challenge it addresses is the *size of data*:
3/
- very large images
- but large images can generate a huge amount of annotations too.
Mastodon can harness these 2 challenges, by staying *interactive and responsive*. You can still modify a single cell in the middle of a dataset of >500M cells
(img from Katie McDole)
- very large images
- but large images can generate a huge amount of annotations too.
Mastodon can harness these 2 challenges, by staying *interactive and responsive*. You can still modify a single cell in the middle of a dataset of >500M cells
(img from Katie McDole)
4/ Mastodon is built with the love we put in #TrackMate and #MaMuT.
It is built as a platform, so that you can implement your own plugins, tracking algos etc... reusing what is already in Mastodon (I will come back to that last).
It is built as a platform, so that you can implement your own plugins, tracking algos etc... reusing what is already in Mastodon (I will come back to that last).
5/
And there are a lot of user-oriented features to make it able to deal with difficult images and challenging and ambitious scientific projects.
Some quick examples:
And there are a lot of user-oriented features to make it able to deal with difficult images and challenging and ambitious scientific projects.
Some quick examples:
6/
A user interface for fully automated tracking. A bit like #TrackMate, but done better.
And it can process large images on a 'normal' computer (below is my laptop tracking the first 10 timepoints of the MaMuT dataset).
A user interface for fully automated tracking. A bit like #TrackMate, but done better.
And it can process large images on a 'normal' computer (below is my laptop tracking the first 10 timepoints of the MaMuT dataset).
7/
If you know #TrackMate a bit, TrackScheme is there, but animated, smoother, and it can harness a large amount of cells (here 1M on Tobias 2012 MacBook).
If you know #TrackMate a bit, TrackScheme is there, but animated, smoother, and it can harness a large amount of cells (here 1M on Tobias 2012 MacBook).
8/
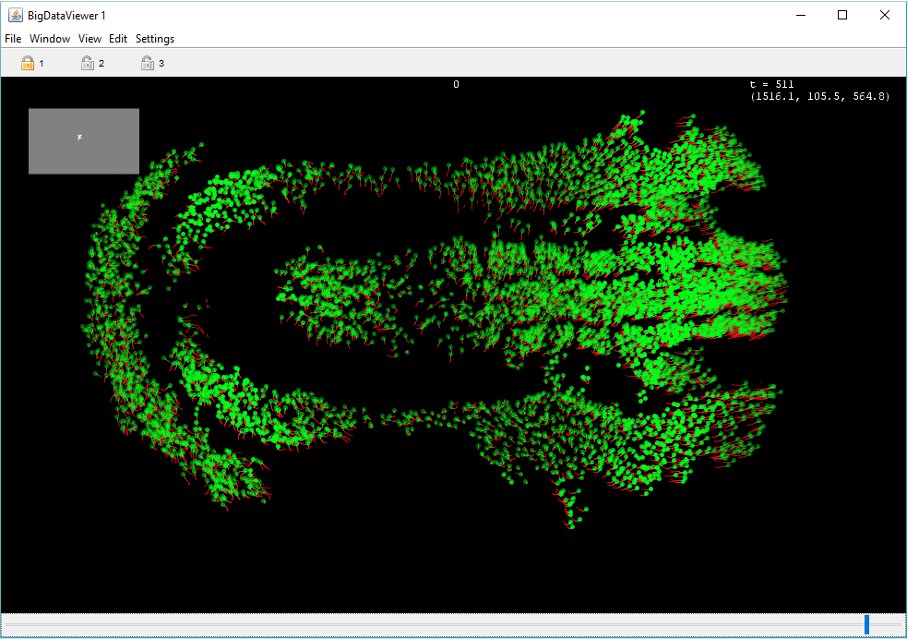
Image visualization if based on the BigDataViewer. A cell is represented as an ellipsoid
Image visualization if based on the BigDataViewer. A cell is represented as an ellipsoid
9/
A lot of effort was made to have sync views, to keep the user oriented in possibly large dataset.
Visual cues with shared selection, highlight and focus over several views
A lot of effort was made to have sync views, to keep the user oriented in possibly large dataset.
Visual cues with shared selection, highlight and focus over several views
10/
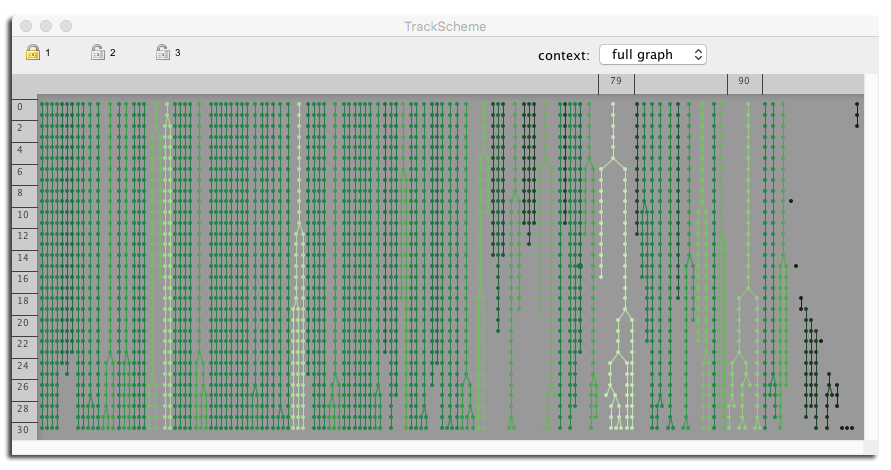
The notion of "spatial context" in TrackScheme: you only see the lineage of what is displayed in another view:
The notion of "spatial context" in TrackScheme: you only see the lineage of what is displayed in another view:
11/
Undo and redo when you are editing manually!
My personally most cherished feature
Undo and redo when you are editing manually!
My personally most cherished feature
12/
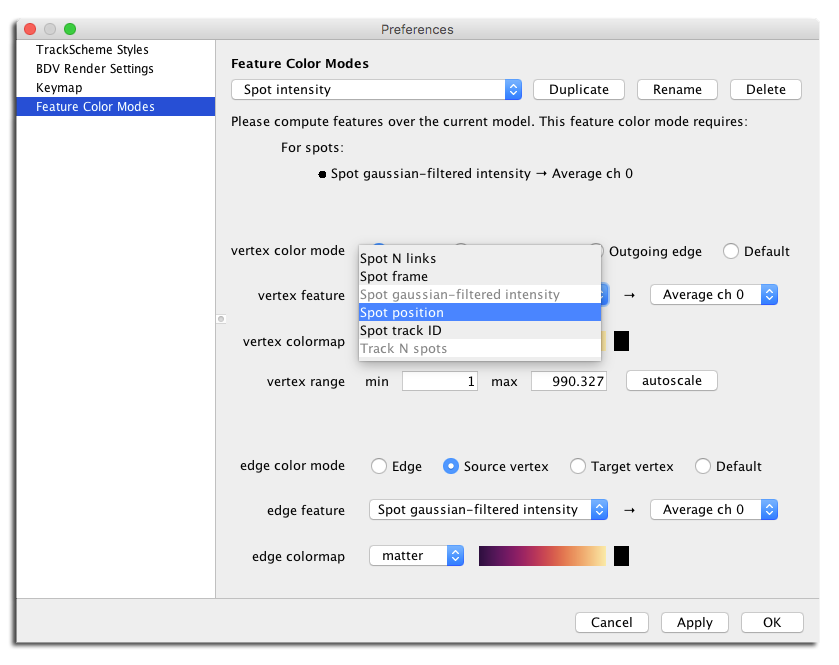
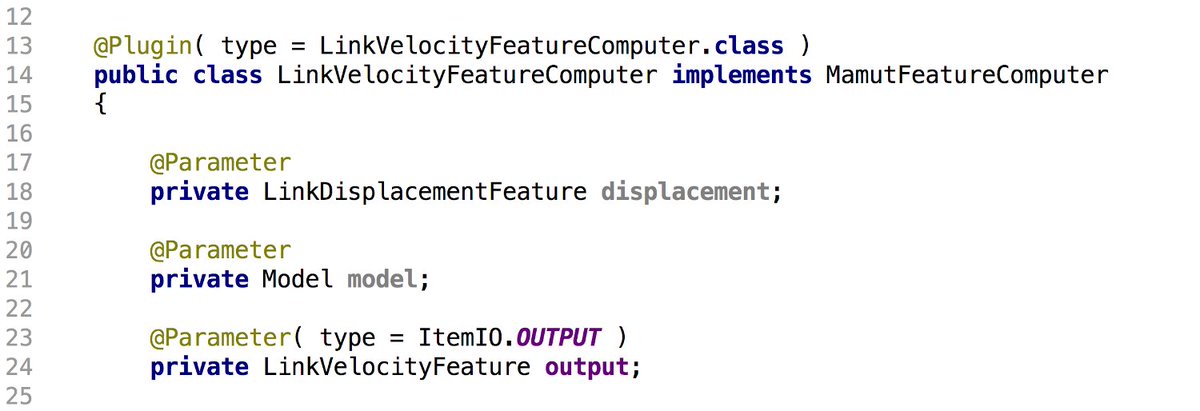
Analysis tools are there to. There is numerical feature mechanism, that you can extend at will
Analysis tools are there to. There is numerical feature mechanism, that you can extend at will
13/
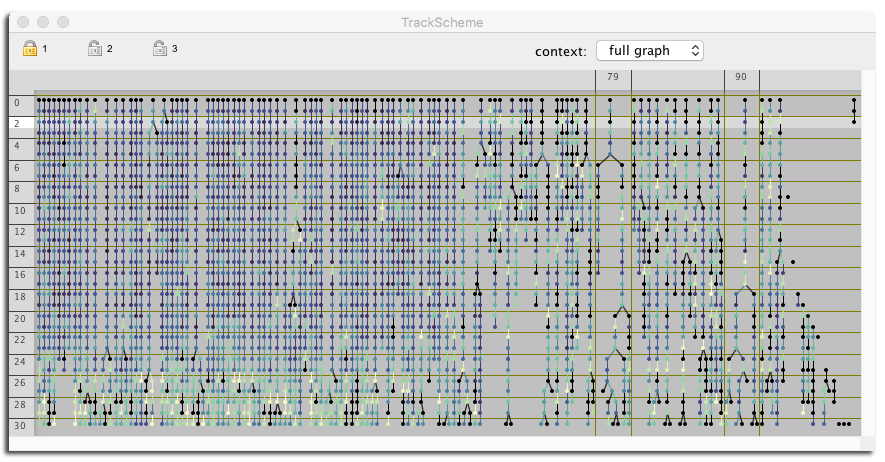
And you can use the feature values to color about everything so that you can detect interesting events in an easier manner:
And you can use the feature values to color about everything so that you can detect interesting events in an easier manner:
14/
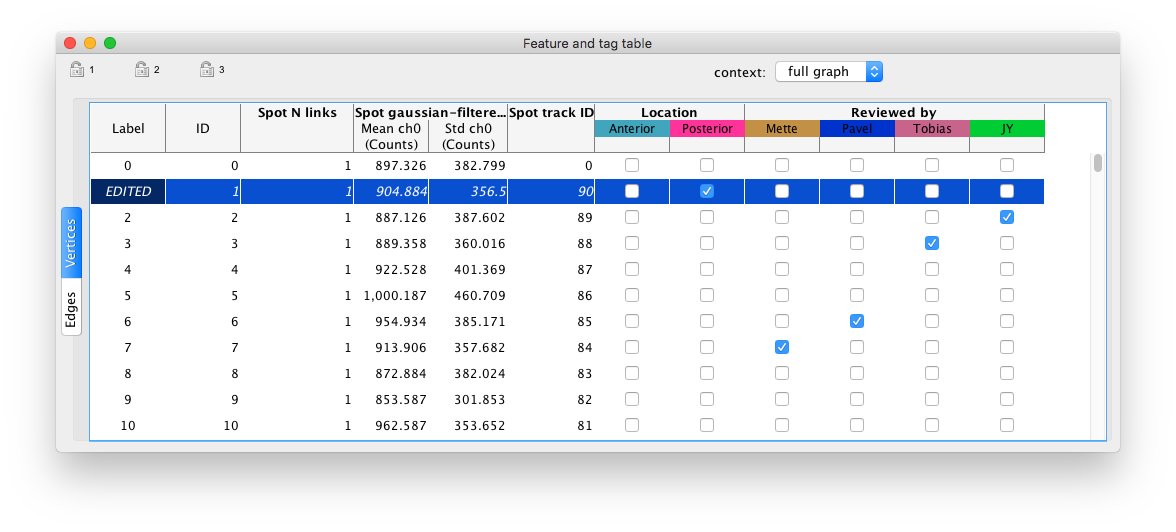
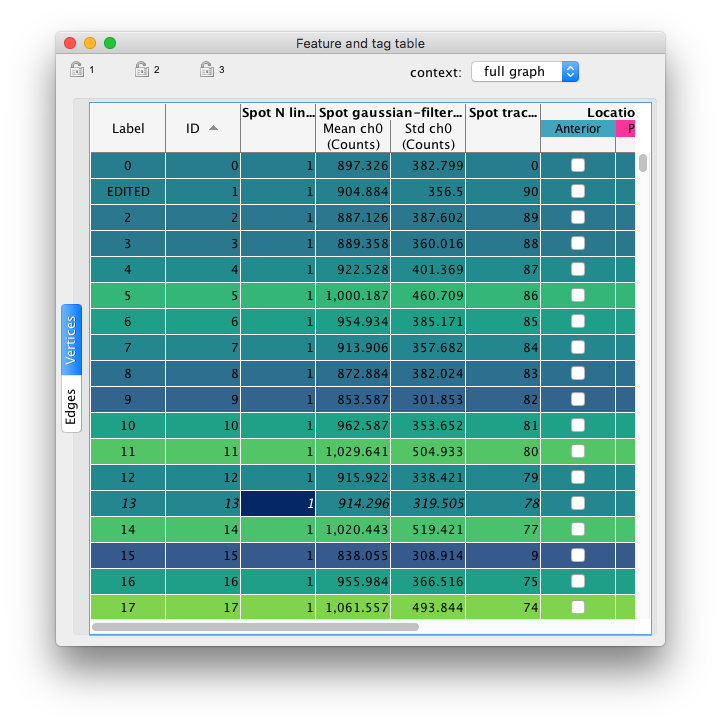
There is also a table view to display and export numerical results.
And tags! you can tag some cells / tracks with whatever you want: anatomical labels, review status etc...
There is also a table view to display and export numerical results.
And tags! you can tag some cells / tracks with whatever you want: anatomical labels, review status etc...
15/
Tags can be edited in the view or in the table:
Tags can be edited in the view or in the table:
16/
And Mastodon is YOUR software: It is fully extensible, whether you want to add an analysis feature, a new tracking algorithm or a misc plugin, you can (thanks #SciJava!)
And Mastodon is YOUR software: It is fully extensible, whether you want to add an analysis feature, a new tracking algorithm or a misc plugin, you can (thanks #SciJava!)
17/
We are super thrilled about what is Mastodon developing into. Now it is mature enough to be used by others and a beta (understand: probably full of bugs) is available.
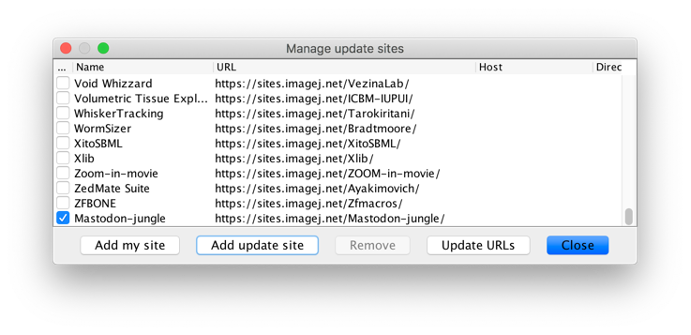
You can get it via Fiji update site mechanism for now:
(add https://sites.imagej.net/Mastodon-jungle manually)
We are super thrilled about what is Mastodon developing into. Now it is mature enough to be used by others and a beta (understand: probably full of bugs) is available.
You can get it via Fiji update site mechanism for now:
(add https://sites.imagej.net/Mastodon-jungle manually)
18/
I get to make presentations and tweets about Mastodon, but the reason I say it is fantastic is because Tobias did most of the job
We had to design our own data model from scratch and @pietzscht was the lead and the main dev.
Also fantastic help from @VladimirUlman
I get to make presentations and tweets about Mastodon, but the reason I say it is fantastic is because Tobias did most of the job

We had to design our own data model from scratch and @pietzscht was the lead and the main dev.
Also fantastic help from @VladimirUlman
19/
Does Mastodon look interesting to you?
Then apply to its tutorial in the I2K conference https://www.janelia.org/you-janelia/conferences/from-images-to-knowledge-with-imagej-friends
Does Mastodon look interesting to you?
Then apply to its tutorial in the I2K conference https://www.janelia.org/you-janelia/conferences/from-images-to-knowledge-with-imagej-friends
20/
Oh yeah, and there is a WIP of a user manual.
https://github.com/mastodon-sc/mastodon-manual/blob/pdf/MastodonManual.pdf
Please excuse the tone I am still working on this.
Oh yeah, and there is a WIP of a user manual.
https://github.com/mastodon-sc/mastodon-manual/blob/pdf/MastodonManual.pdf
Please excuse the tone I am still working on this.

 Read on Twitter
Read on Twitter