Very excited to see our work on deep learning germline variant characterization out today in @JAMA_current
Here is a quick thread on the major findings and takeaways from this study.
#CancerResearch
#ProstateCancer
#Melanoma
1/n
https://jamanetwork.com/journals/jama/fullarticle/10.1001/jama.2020.20457?guestAccessKey=39889aad-2894-4380-b869-5704ed2f9f6b&utm_source=twitter&utm_medium=social_jama&utm_term=4168566540&utm_campaign=article_alert&linkId=104709446
Here is a quick thread on the major findings and takeaways from this study.
#CancerResearch
#ProstateCancer
#Melanoma
1/n
https://jamanetwork.com/journals/jama/fullarticle/10.1001/jama.2020.20457?guestAccessKey=39889aad-2894-4380-b869-5704ed2f9f6b&utm_source=twitter&utm_medium=social_jama&utm_term=4168566540&utm_campaign=article_alert&linkId=104709446
Germline genetic analysis is an integral part of precision cancer medicine with established diagnostic, prognostic & therapeutic utility. However, only a small fraction of cancer patients have identifiable pathogenic variants to explain (at least in part) their cancer risk
2/n
2/n
So in this study, we sought to see if recent advances in machine learning can expand the diagnostic yield of germline analysis and identify additional cancer patients with *clinically informative* pathogenic variants that escaped detection using standard analysis approaches
3/n
3/n
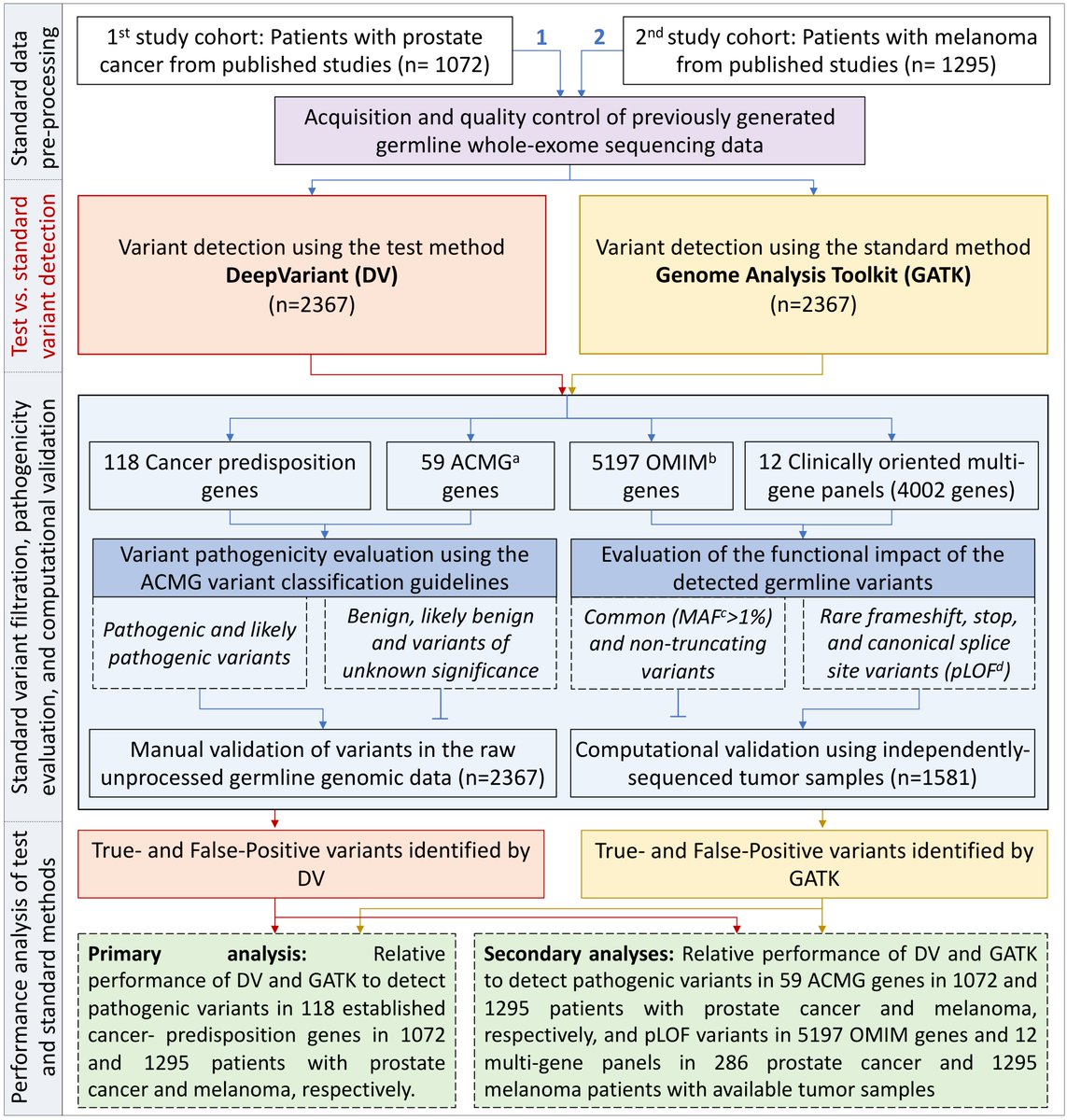
To explore this, we analyzed the germline WES data of 2,367 patients with prostate cancer or melanoma twice, once using DeepVariant (a deep neural network analysis framework by @GoogleAI) and then using GATK (the most widely used “standard” germline variant detection method)
4/n
4/n
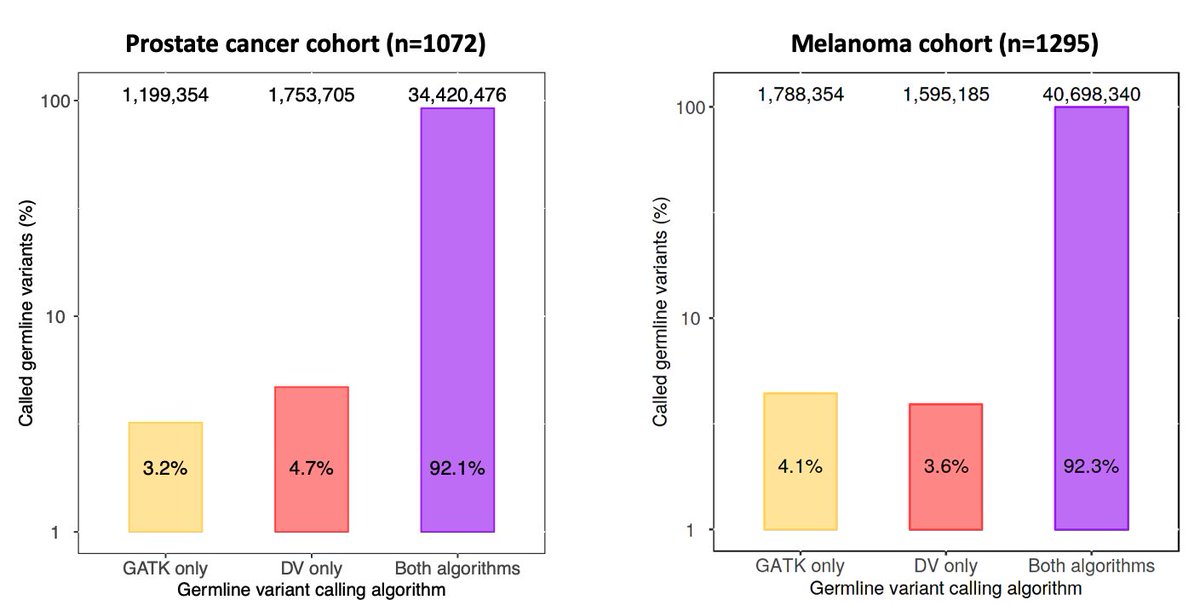
Overall, approximately 8% of all rare and common germline variants (total ~80 million) were only detected by one method but not the other, suggesting a significant discrepancy between these variant detection approaches.
5/n
5/n
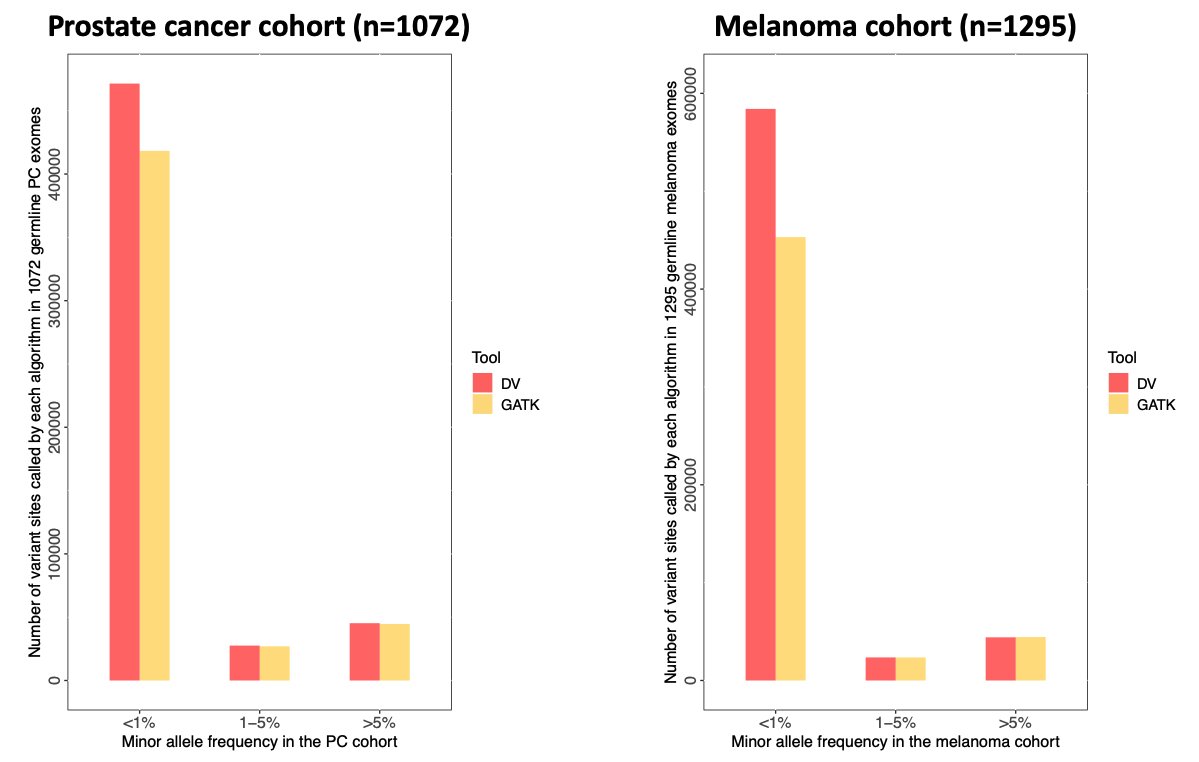
Our analysis also showed that while the deep learning and the standard methods had a comparable performance for common germline variants (MAF>1%), deep learning called many more rare variants (MAF<1%) across all examined cancer cohorts (PC and melanoma)
6/n
6/n
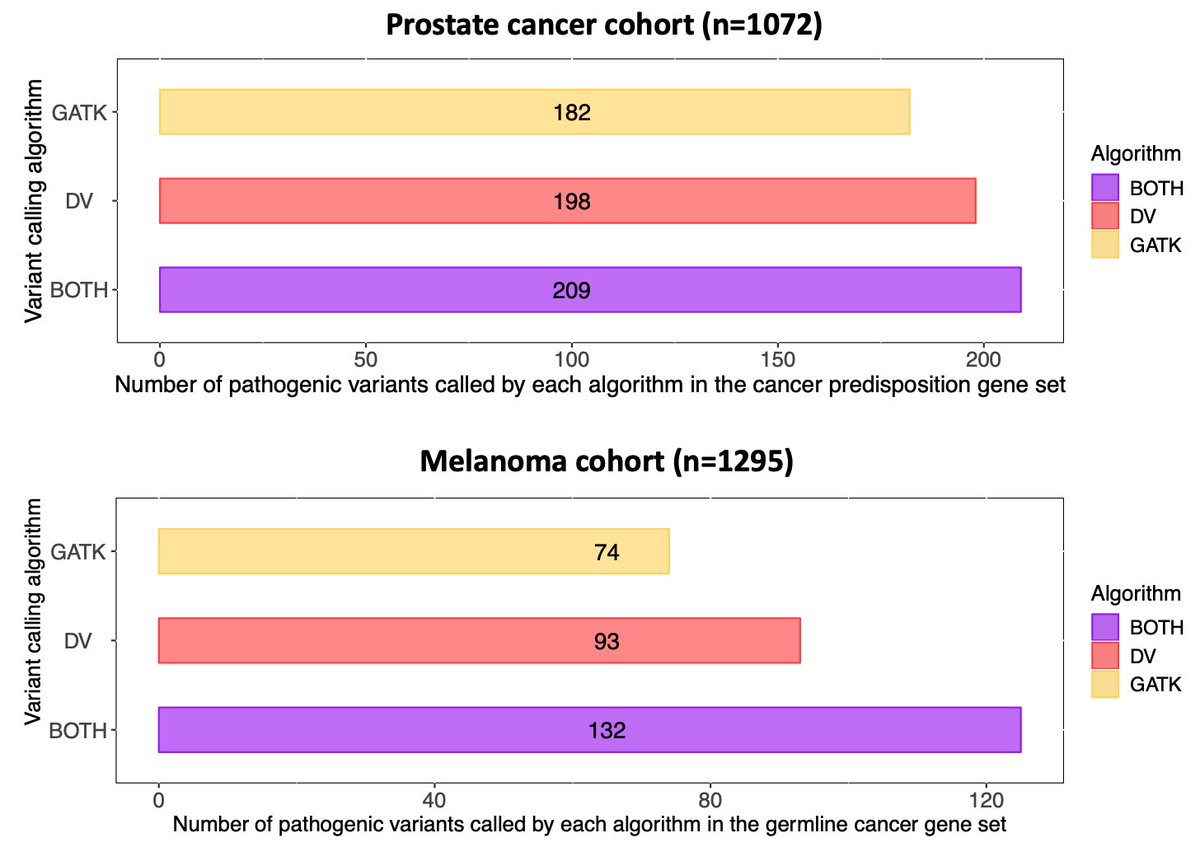
When looking at the cancer-predisposition genes, deep learning identified 16 *additional* PC patients and 9 *additional* melanoma patients with informative pathogenic variants that went undetected by the standard method resulting in higher sensitivity/specificity/PPV/NPV
7/n
7/n
Some of the validated pathogenic cancer risk variants exclusively detected by deep learning in the cancer-predisposition genes (such as the ones highlighted here) have established gene-specific recommendations for cancer prevention and/or cancer treatment
8/n
8/n
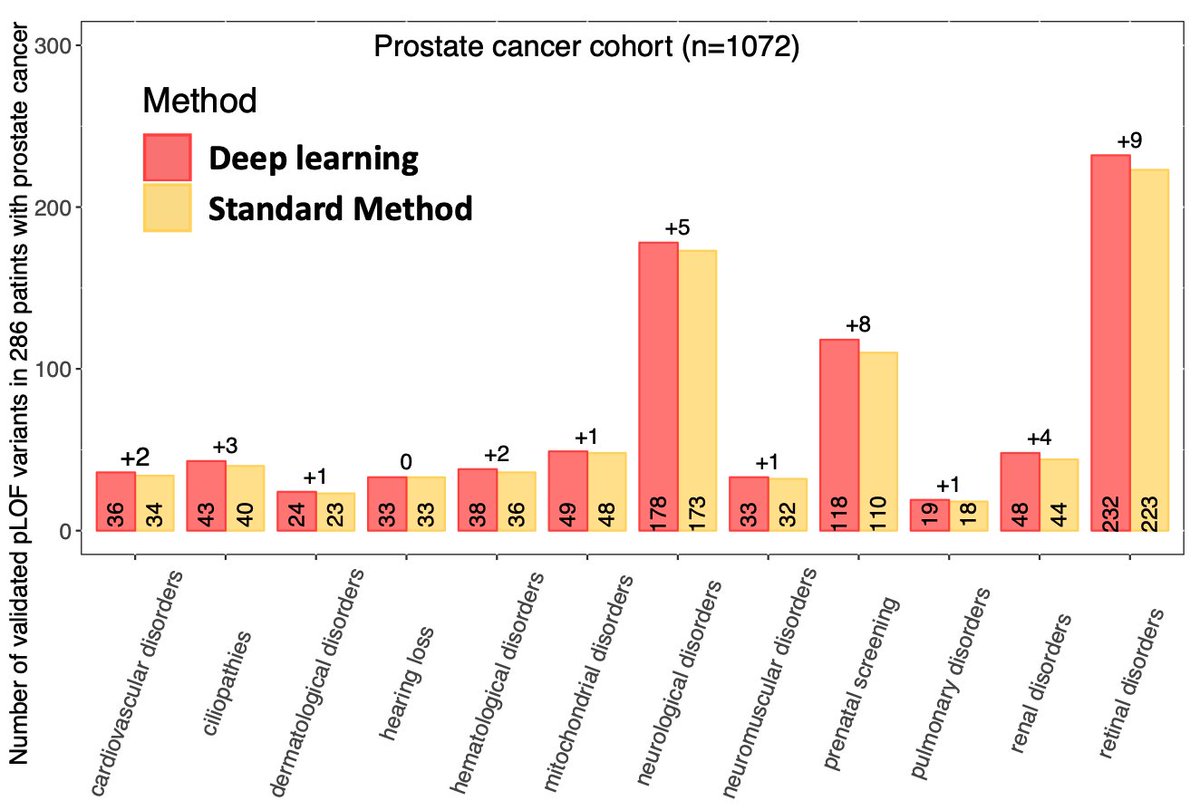
Similarly, deep learning detected more germline pathogenic and LOF variants in 59 highly penetrant ACMG genes, 5197 clinically relevant mendelian genes in OMIM, and 12 clinically-focused gene panels used to diagnose various non-cancer phenotypes.
9/n
9/n
Collectively, this study showed that deep learning can enhance the diagnostic yield of germline analysis and expand the utility of this tool to additional patient groups who may not otherwise be eligible for certain preventive and/or therapeutic interventions
10/n
10/n
Importantly,deep learning did not identify all pathogenic variants that existed in the raw genomic data so it is not perfect. Also this study was conducted under a specific set of settings (listed in Limitations) which should be kept in mind when interpreting the findings
11/n
11/n
Overall, this study also highlights the potential to further enhance novel genetic discoveries in the germline space when using deep learning to augment the established germline variant approaches
12/n
12/n
Finally, special thanks to @VanAllenLab and all collaborators for making this a successful team effort. Also, grateful to the Prostate Cancer Foundation @PCFnews/ @PCF_Science and the ASCO Conquer Cancer Foundation @ASCO/ @ASCOPost for supporting my research
13/13
13/13

 Read on Twitter
Read on Twitter