Excited to share FREE final PDF of our article with the amazing @mkbaine on SCLC subtypes https://authors.elsevier.com/a/1c0H65Xq2QKdvX @JTOonline. Thanks again to the entire team @charlesrudin @triparnasen @wvickylai & @MSKPathology!
So… is this ready for prime time & what did we learn? 1/
1/
So… is this ready for prime time & what did we learn?
 1/
1/
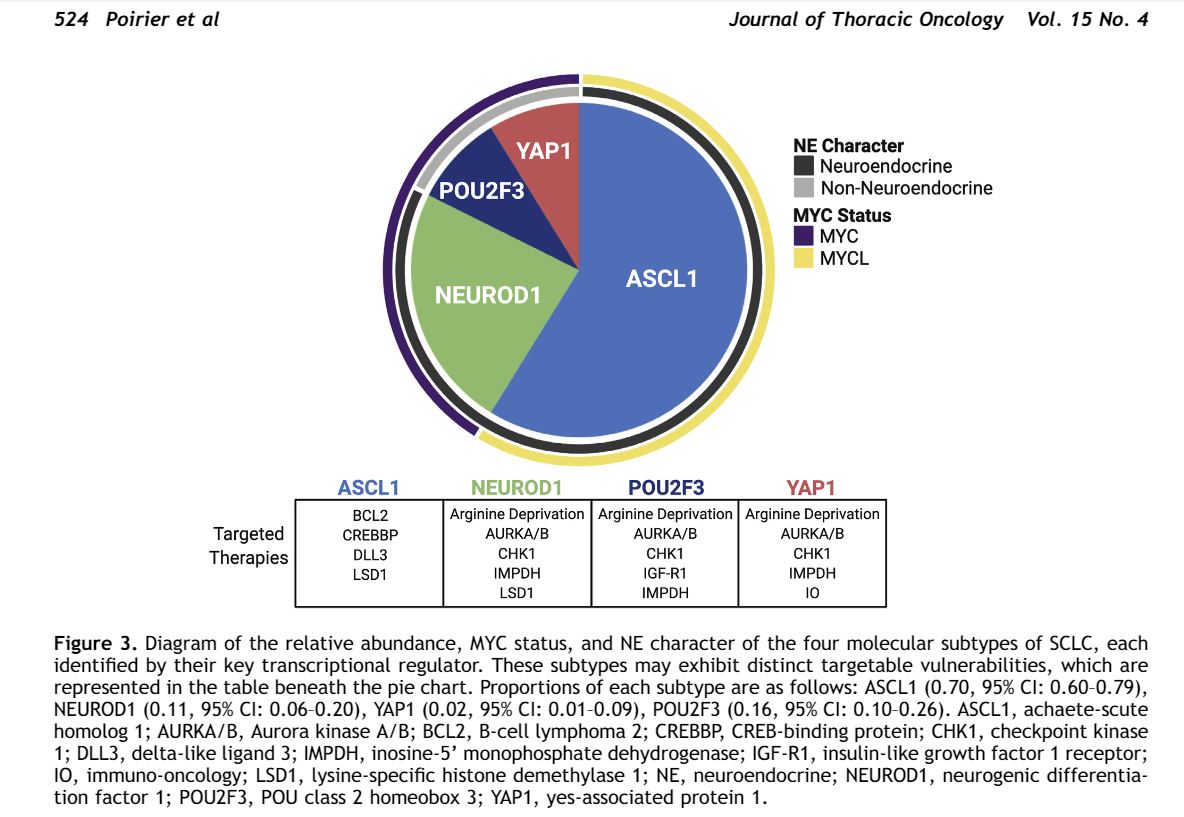
So…ready for primetime? NO. Biomarker-based Tx of SCLC remains a pipedream… for now! A lot of work from SCLC researchers predicts different responses to various therapies in transcriptional subtypes of SCLC based on expression of these 4 markers: 2/
Our study shows that it is feasible to identify these subtypes by IHC in clinical samples, but clinical utility still remains TBD. Also exact correspondence of some subtypes predicted by mRNA vs IHC will require clarification. 3/
What DID we learn? (for SCLC aficionados):
-similar distribution of subtypes to what is predicted in the models, BUT…
-MUCH more co-expression of 2 major markers (ASCL1 & NEUROD1)
-no clear YAP1 group
-confirmation of VERY interesting POU2F3 (tuft cell marker)+ group
4/
-similar distribution of subtypes to what is predicted in the models, BUT…
-MUCH more co-expression of 2 major markers (ASCL1 & NEUROD1)
-no clear YAP1 group
-confirmation of VERY interesting POU2F3 (tuft cell marker)+ group
4/
Also for SCLC aficionados - pl see our paper for a deep dive into comparison of usual SCLC markers and histology in transcription factor-based subtypes 5/
Hoping this study will be a small step in bringing all the exciting progress from SCLC research community closer to clinical practice. Thanks for all your amazing work! @TGOliver2 @christine_lovly @AnnaFaragoMD @ChrisVakoc
Thanks everyone for your attention! 6/
Thanks everyone for your attention! 6/

 Read on Twitter
Read on Twitter