THREAD: Does bioinformatics need fundamentally different compute platforms? Lots of thoughtful innovation, but adoption is low. Need to balance solving a niche pain point with a domain specific solution, vs using a more general tool. Some musings sparked by 3 recent papers: [/1]
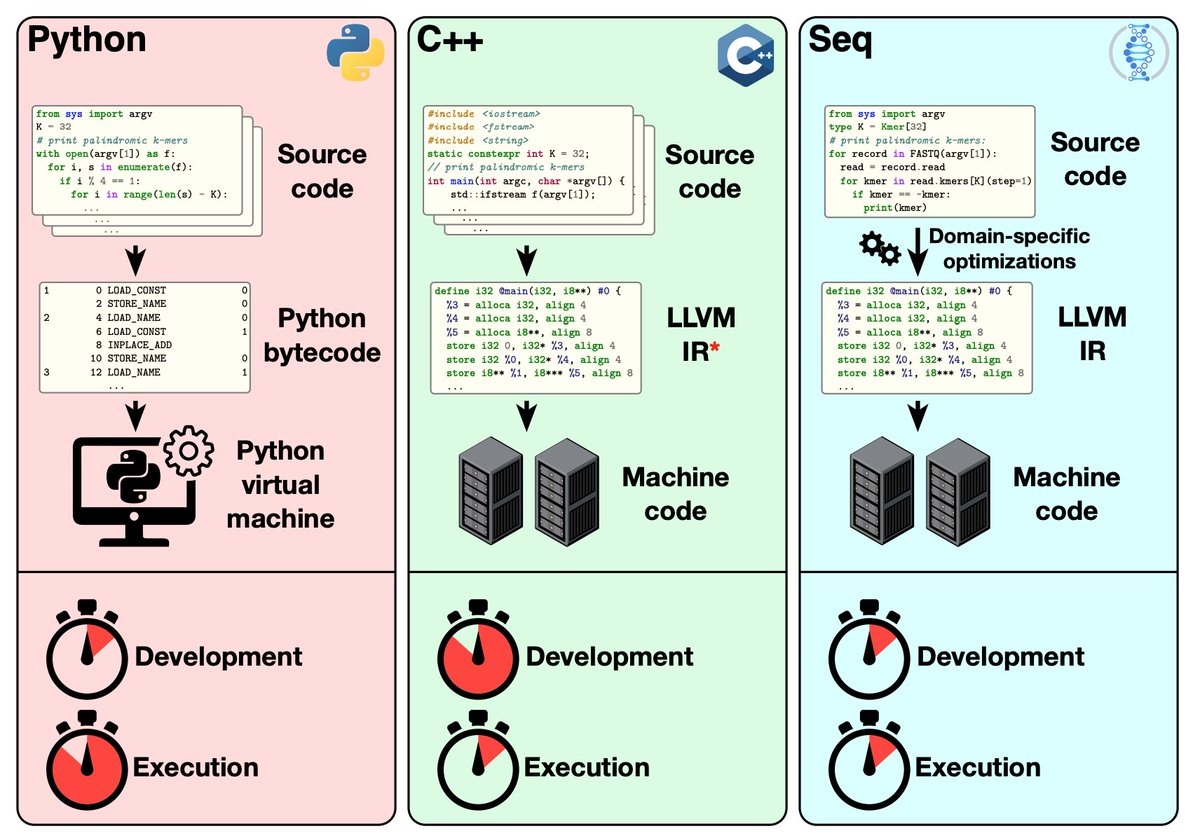
(1) Language: Numpy is not all you need. Common sequence data computations can be optimized with specialized tools, and there’s tons of niche bioinformatics packages, but I was blown away by Seq: a new programming *language* (+ compiler) for bioinformatics from Berger lab@MIT /2
Seq has python syntax (actually its a statically-typed subset of Python) but is LLVM backend so has C-like performance. Treats nucleotide sequence, kmers, and piping as 1st class citizens. Super impressive: common bioinf. algorithms are elegant and readable but also fast! /3
“Seq is to bioinformatics what MATLAB is to numerical computing”. But switching to a domain specific language, even one that runs most native python, is a giant barrier. (Once I left Matlab for python I never looked back) https://www.biorxiv.org/content/10.1101/2020.10.29.361402v1 https://dl.acm.org/doi/10.1145/3360551 /4
(2) Storage: managing sequencing data is painful - large files need compression, but many bioinf tools require uncompressed/derivative formats/random access. LOTS happening in compression methods ( https://bit.ly/35zXtFY ) but I was impressed by clever file system tool FASTAFS: /5
Its a virtual file system (using FUSE) that stores compressed sequencing data but allows decompressed random access in form of virtual files. Backward compatible with tools that are expecting uncompressed fasta files without having to store uncompressed data. /6
Cool idea and FUSE is awesome but overhead/performance hard to predict at scale in production. https://www.biorxiv.org/content/10.1101/2020.11.11.377689v1.full.pdf /7
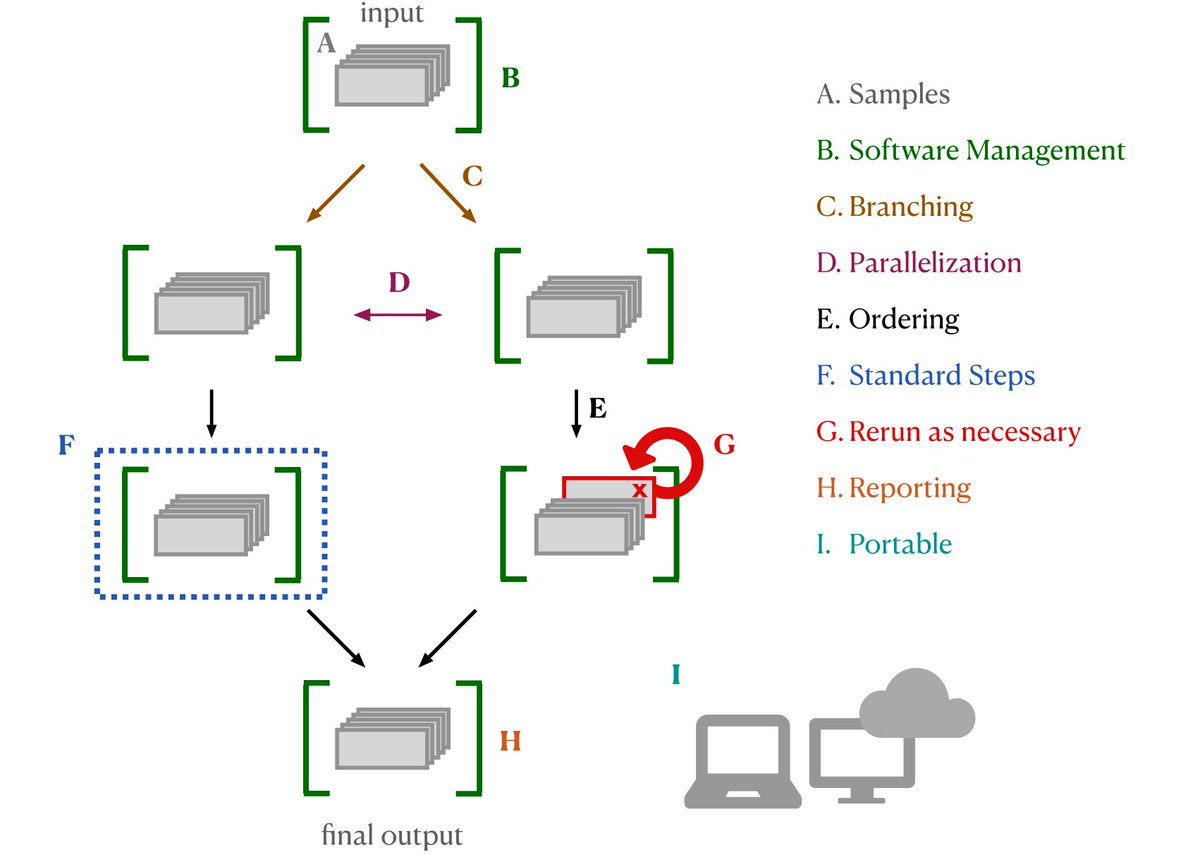
(3) Workflows: I would be remiss not to mention the workflow automation war! If you’re in the field you’ve def encountered these- Nextflow/Snakemake/Reflow/wdl/CWL are all designed to shepherd seq data thru bioinformatics pipelines. Nice review: https://www.biorxiv.org/content/10.1101/2020.06.30.178673v2 /8
But there are similar tools in ETL/Data Science world, where Airflow is still top dog and Prefect/Metaflow/Argo/++ are muscling in, and can be used for bioinf too (with tradeoffs). Caching, data provenance, introspection, backend compatibility, scalability - what do you need? /9
The number of options is mind boggling: https://github.com/pditommaso/awesome-pipeline /10
Final thoughts on how we think ab this at DZD: as a genomics startup you betcha we use plenty of open source bioinformatics tools. We have to balance maintainability, performance, and ease of use (which differs b/w bioinformaticians, data scientists, and software engineers). /11
SO unsurprisingly the answer to when we use a domain specific tool is “it depends” :). [/fin]

 Read on Twitter
Read on Twitter