A short thread on this interesting new paper.
Transferring polygenic risk scores (PRSs) across populations is one of the most urgent problems in the field. The idea presented here is perhaps not currently practical but is still thought provoking.
1/8 https://www.biorxiv.org/content/10.1101/2020.11.12.373647v1
Transferring polygenic risk scores (PRSs) across populations is one of the most urgent problems in the field. The idea presented here is perhaps not currently practical but is still thought provoking.
1/8 https://www.biorxiv.org/content/10.1101/2020.11.12.373647v1
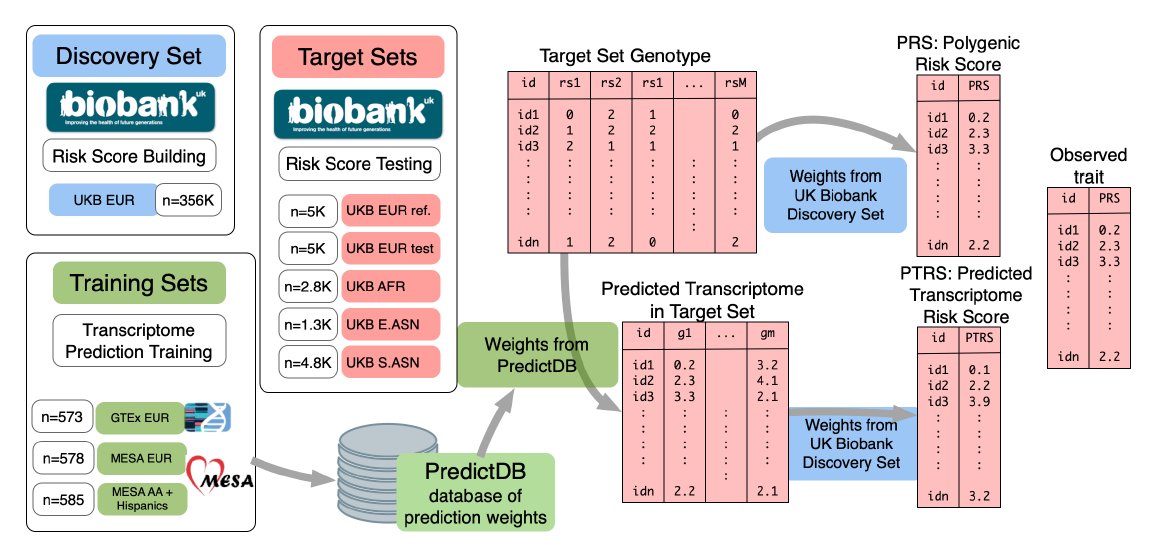
The framework is explained in the diagram. (1) Download existing models from GTEx/similar datasets that predict gene expression based on genetic variants. (2) Take a large sample ("discovery" set) with genotypes/phenotypes. Use it to compute standard PRS based on
2/8
2/8
genotype/phenotype association. (3) In parallel, predict gene expression level for each gene and each individual in the discovery set. Use elastic nets to estimate the effect size of (the expression of) each gene on the phenotype (still in the discovery set).
3/8
3/8
(4) In a "target" set, again predict the expression of each gene in each individual. (5) Also in the target set, compute standard PRS, as well as "transcription" PRS (PTRS), which is the sum of predicted expressions of genes, each gene weighted by the effect size learned
4/8
4/8
in the discovery set. (6) Finally, compare the accuracy of prediction between PRS and PTRS, and evaluate how both deteriorate across populations. For PTRS, compare accuracy when using gene expression prediction models learned in different populations.
5/8
5/8
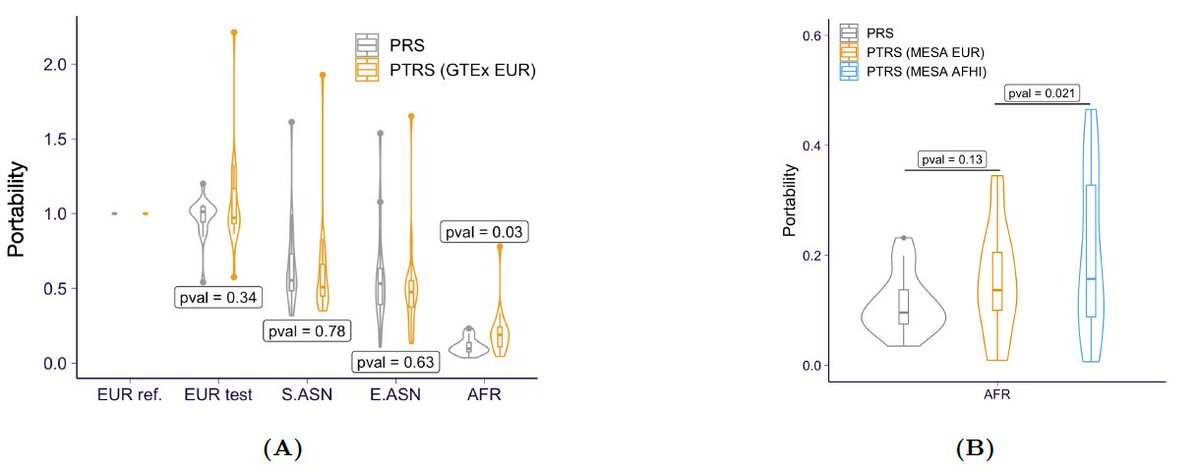
The motivation is that (1) effect sizes of genes should be more stable across populations than genetic effects (which depend on complicated LD patterns); and (2) that gene expression models can be accurately learned in non-European populations even using small samples.
6/8
6/8
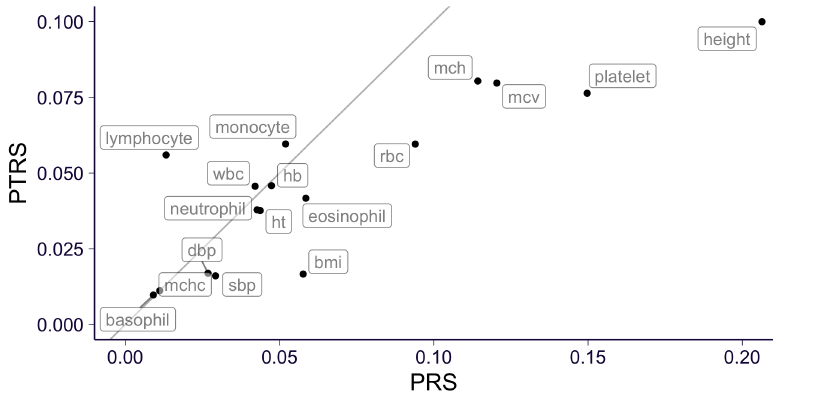
They show that PTRS performs worse than PRS for most traits. However, the decrease in the accuracy of the PTRS in Africans is smaller than that of the PRS, and is even smaller when the expression model was learned in Africans.
7/8
7/8
The limitations are (1) expression was predicted based on a single tissue (and not necessarily the most relevant one); and (2) that it would obviously be better to use genotypes + predicted expression jointly. But it is still a very interesting approach.
8/8
8/8

 Read on Twitter
Read on Twitter