(1/6) Remember how I was stuck in the US due to covid, after visiting the statistical genetics workshop in Boulder? Well, worth it! A lecture of Mike Hunter on multilevel twin models very much inspired me & @SofiekeKevenaar, resulting in our first paper https://www.biorxiv.org/content/10.1101/2020.11.11.377820v1
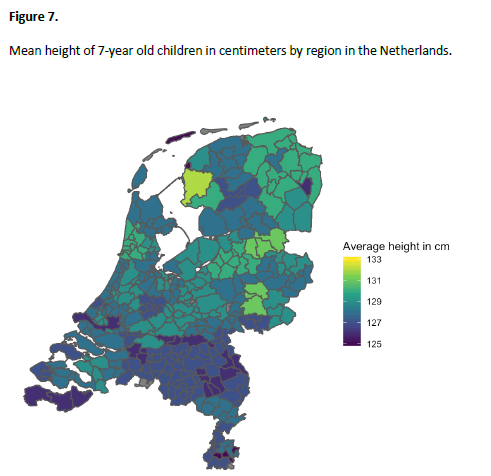
(2/6) We illustrated the use of the ML twin model by fitting it to @NTRscience data of children's height. The cool thing about the ML approach? It gave us a quick and easy way to investigate clustering of height in regions of the NL and also correct for genetic ancestry.
(3/6) We found that region explains 1.8% of the variance in children's height. When unmodelled, the region effect would have been captured by the C-component. The variance explained by region was 7% of this initial common environmental variance.
(4/6) However, in a subsample of participants with genome-wide SNP data, the effect of region was explained away when we included the first genetic principal component in the model.
(5/6) Our results suggest that the phenotypic variance explained by region actually represent ancestry effects on height! 


 Read on Twitter
Read on Twitter