Hey chromatin folks! I am incredibly excited to share my first preprint paper and first paper from the Funabiki lab with you! "Near-atomic resolution nucleosome structures and their variations in interphase and metaphase chromosomes" https://www.biorxiv.org/content/10.1101/2020.11.12.380386v1
We determined the near-atomic resolution structure of nucleosome in interphase and metaphase chromosome by combining the Xenopus egg extract system I have learned at the @HironoriFunabi1 and the nucleosome structural biology I have learned at the @Kurimizakalab.
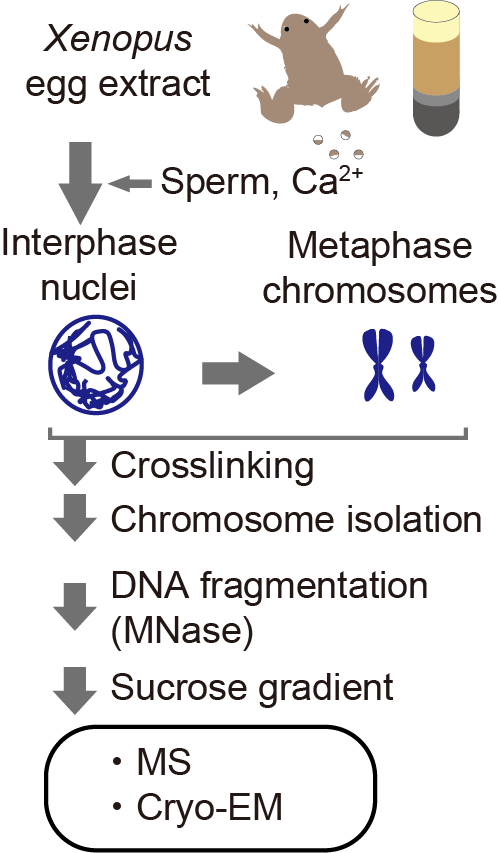
Using Xenopus egg extract, we made functional interphase and metaphase chromosomes on the Xenopus genome DNA and isolated the crosslinked interphase and metaphase chromosomes with the exact identical procedure. Then determined the nucleosome structures at up to 3.4Å resolution.
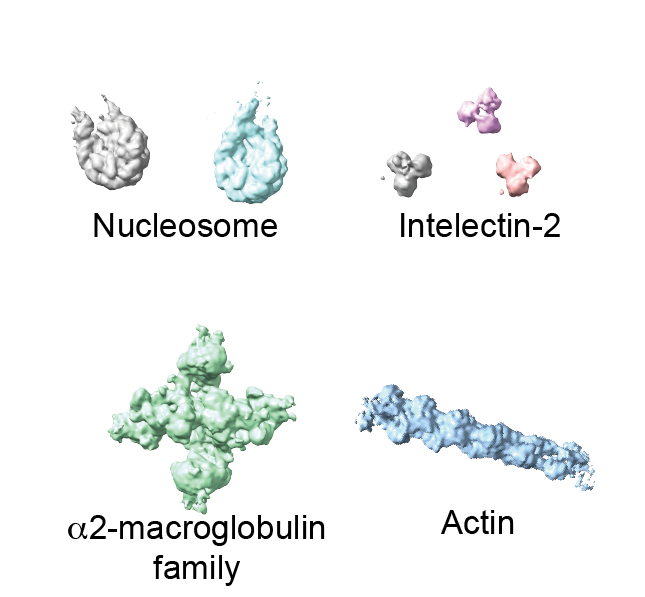
With tremendous help from Seth Darst lab and @RubyFroom in his lab, we determined the structures of the five most abundant complexes (nucleosome, chromatosome, alpha2-macroglobulin, intelectin-2, actin filament) at once in an unbiased way (Fig. 2).
Since alpha2-macroglobulin is the extracellular protein, we have never expected that we determine this protein's structure. Interestingly, the structure of "native" alpha2-macroglobulin was dissimilar to the crystal structure of the recombinant protein.
(if you have a friend working on alpha2-macroglobulin, please let them know that the "native" alpha2-macroglobulin structure (Fig S3G) is available now. )
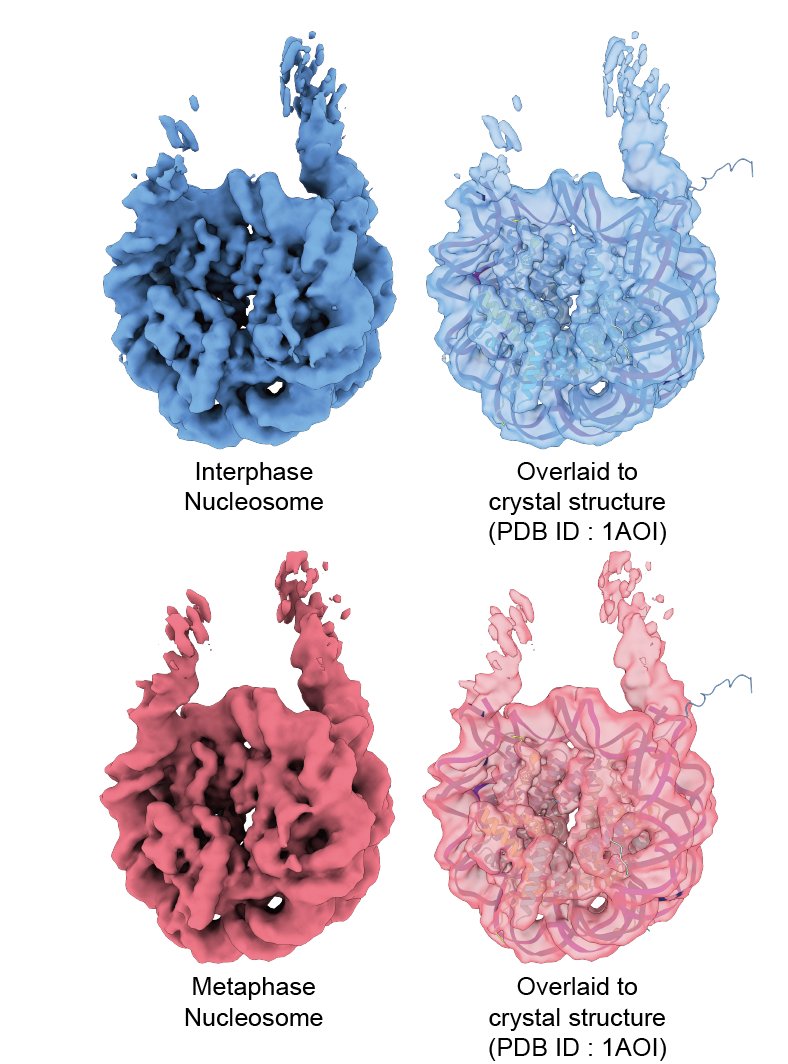
Unlike alpha2-macroglobulin, the average structures of "in chromosome" nucleosomes were perfectly matched with the crystal structure of the nucleosome (Fig. 2, 3). (Probably a good news for nucleosome folks! )
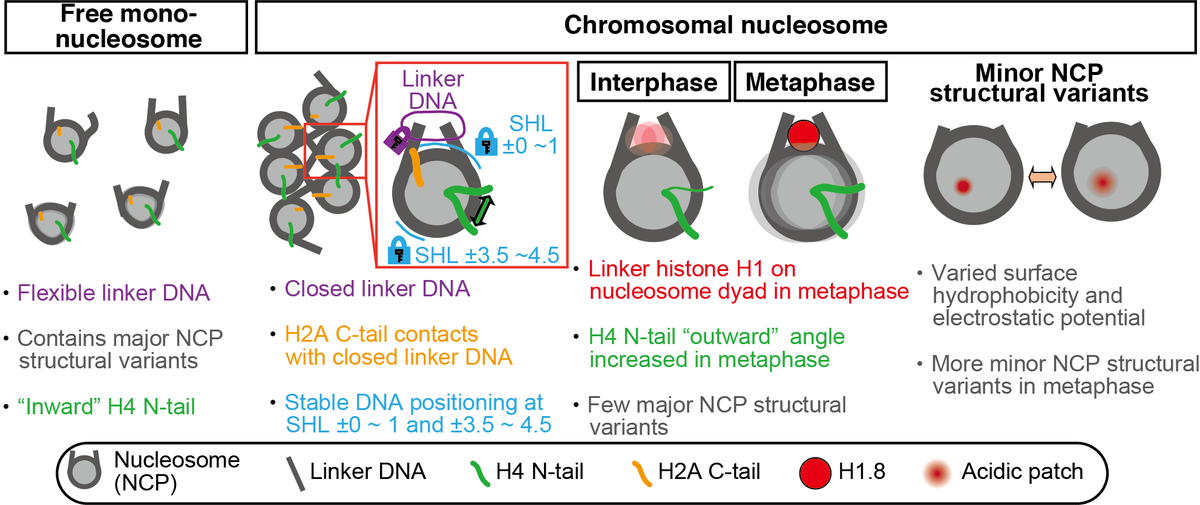
Then, to understand the structural variety of the "in chromosome" nucleosome, we used the 3DVA of cryoSPARC (Fig. 4).
Compared to the free mono nucleosome, both interphase and metaphase nucleosomes had more closed linker DNA and stable nucleosome core particle structure (Fig. 4). Our results suggest that there is an active mechanism to close the linker DNA of nucleosomes in chromosomes (Fig. 5)
Unexpectedly, exclusively for metaphase nucleosome, we found the linker histone H1.8 at the on-dyad position (Fig 4) and determined the H1.8 bound nucleosome structure at 4.4 Å resolution (Fig. 6).
Further 3DVA analysis revealed two types of nucleosome core particle (NCP) structure variants (Fig. 4).
One is the "major structural variants," which are characterized by jagged NCP outlines with sliding of the H3-H4 tetramer, as compared to the smoother circular shape of the canonical NCP. Our data suggest that this type of movement is suppressed in the chromosome (Fig. 7).
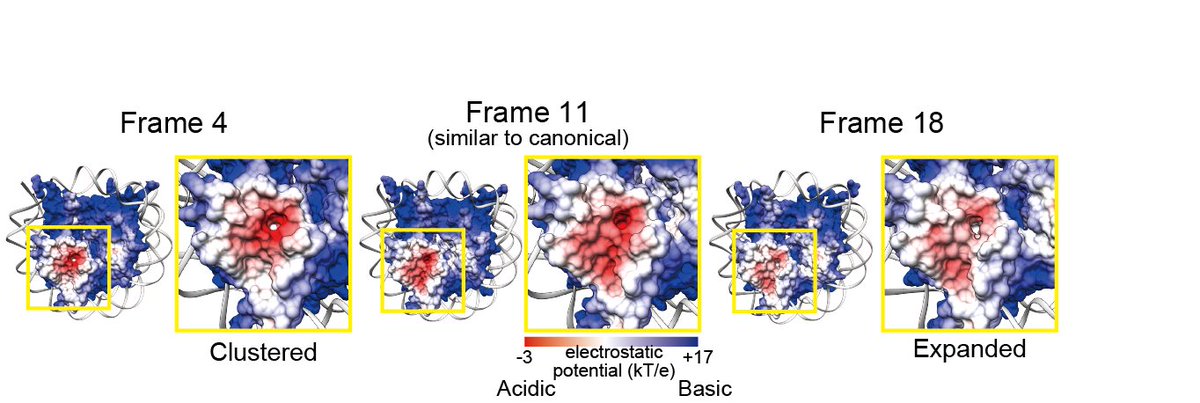
Another is the "minor NCP structural variants," characterized by rotation and reorientation of the H4 N-terminal tail α-helices. Our data suggest that minor NCP structural variants commonly exist in chromosomes and are more prevalent in metaphase than interphase (Fig. 7)
In minor NCP structural variants, H4 N-terminal tail orientation, surface hydrophobicity, and electrostatic potential are changed. These changes may affect the inter-nucleosome interaction and protein binding on nucleosomes and contribute to chromatin compaction in the M phase.
Up until now, no method has been established to analyze the high-resolution nucleosome structure in chromosomes, and the function of the nucleosome structure and its variant in chromosomes can not be addressed.
We hope this study become the first step to link the function and structure of the nucleosome in the chromosome.

 Read on Twitter
Read on Twitter