Delighted to see our latest paper now available on biorxiv:
https://www.biorxiv.org/content/10.1101/2020.11.12.380378v1
In this study, we set out to solve the problem of SNP calling in bacterial pan-genomes. What problem, you say? .... 1/n
https://www.biorxiv.org/content/10.1101/2020.11.12.380378v1
In this study, we set out to solve the problem of SNP calling in bacterial pan-genomes. What problem, you say? .... 1/n
What problem, you say? Roughly speaking, given a set of bacterial genomes, there has been no way to detect all the SNPs between them, including accessory genes. We solve this.
First i'll formulate the problem, give a long bioinformatic discussion, and at the end show some amazing figures/results for E coli. Feel free to skip to the figures 3/n
The problem is this. If you line up two human genomes, the alignable proportion will be 99% or something huge. So, you can go a long way mapping reads to the human reference, and doing SNP calling that way. Not so for bacteria, where that 99% figure could be just 50%. 4/n
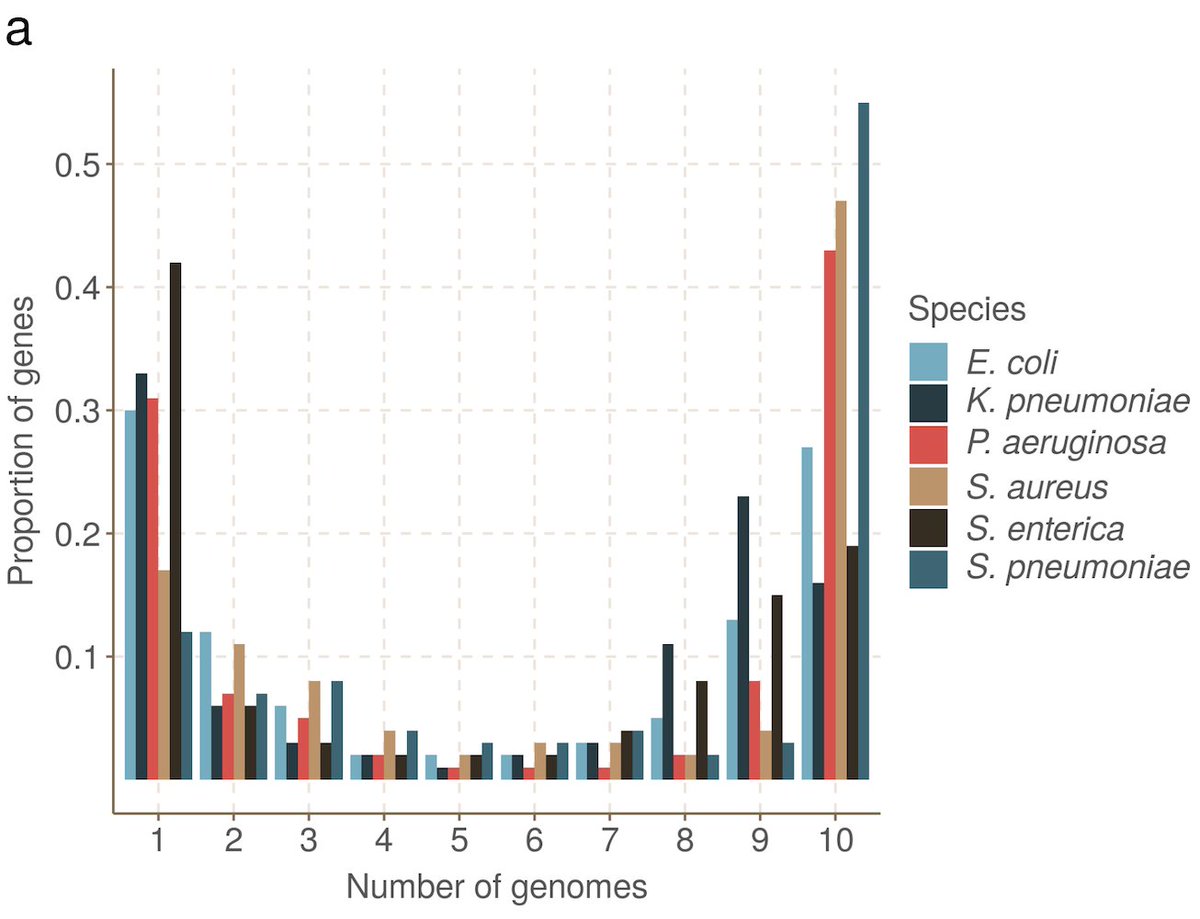
Bacterial genomes are gene-rich (>80%), and follow a universal U-shaped gene frequency distribution - genes tend to be rare or common. There's a core genome shared by most, and a pool or rare and transient genes moving in and out of the population. 5/n
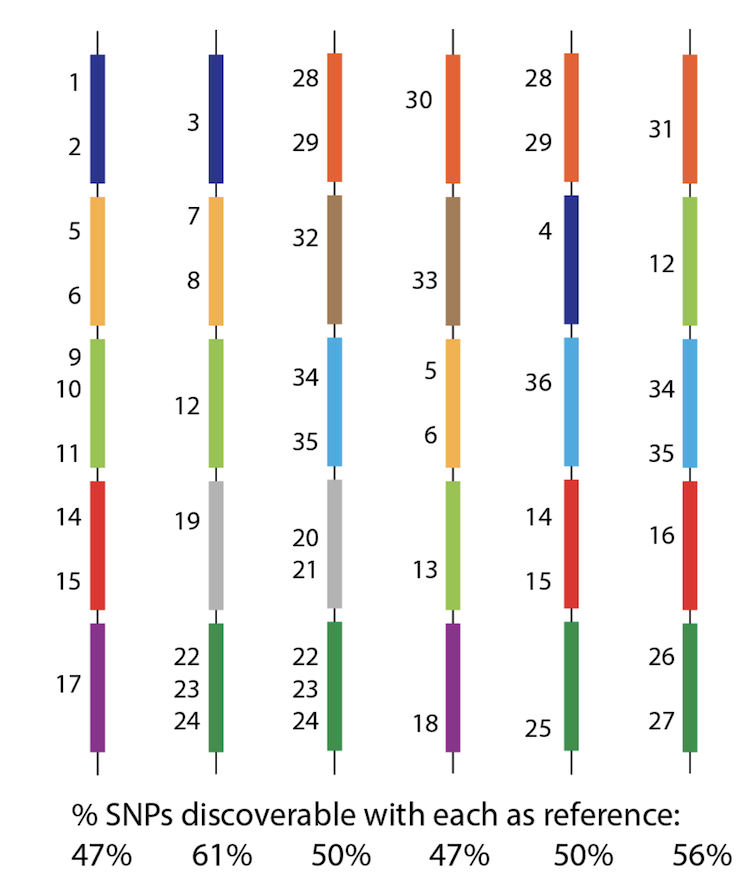
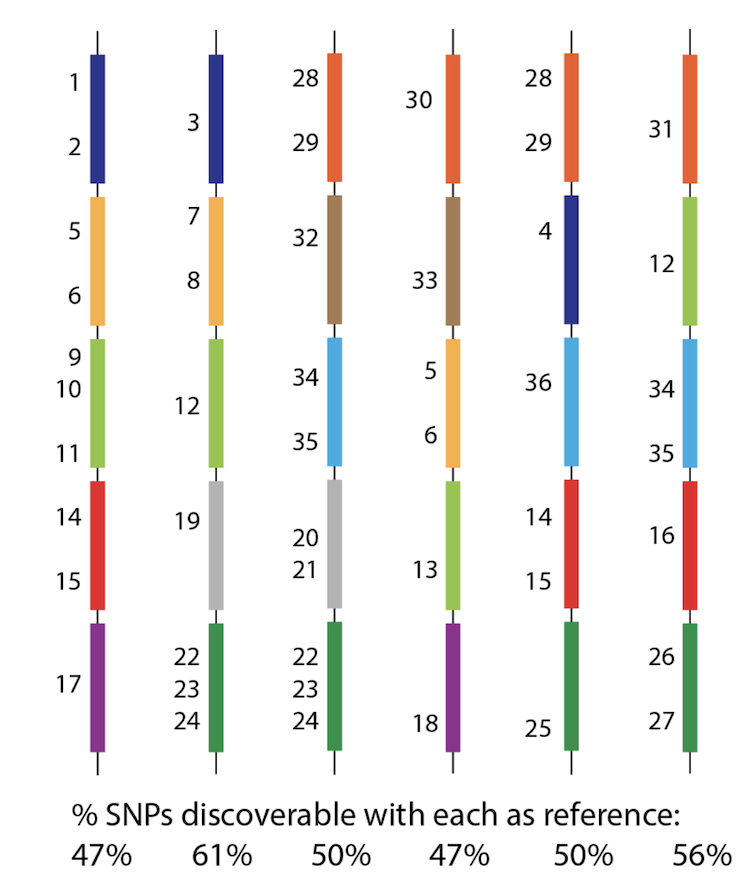
So - how can we possibly call SNPs in a situation like this? Whichever genome we use as reference will be missing many many genes, and therefore SNPs from the population? See this figure, with 36 SNPs, but no reference can see them all. 6/n
attaching end of thread! https://twitter.com/ZaminIqbal/status/1327134512862679043?s=20

 Read on Twitter
Read on Twitter