I've spoken highly of napari here before, and as I just finished training one of my interns on our new napari based tool, here's a short thread about how using @napari_imaging has helped distribute my python bioimage analysis workflows to other members of my lab.
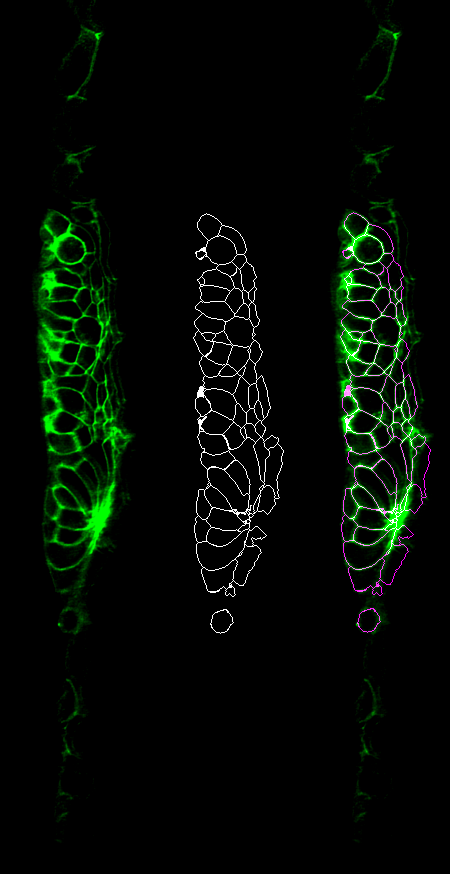
A common first step for us is instance segmentation of cells in our tissue. The segmentation itself is pretty straightforward, and runs in python using jupyter notebooks. And it works pretty well -- here is original, segmented and overlay. But, there are problems.
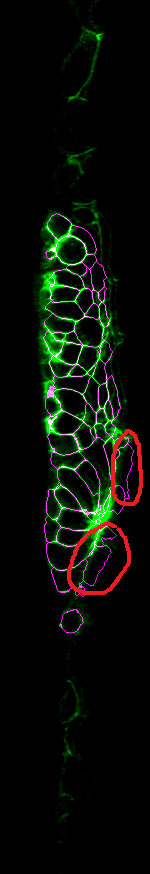
The first problem is that some parts of the background or surrounding tissue get segmented, for instance the circled parts here. Also we sometimes miss or fuse cells in the internal structure of the tissue.
By and large, this is due to a failure in our marker generation for the watershed. We're using a mixture of nuclei and distance transforms on the cells themselves, which works for 95-99% of cells, but not for 100%.
And that leads us to the second problem. We can fix some of these cells programmatically, but some will need to be manually fixed. There's still no substitute for an expert human eye.
Easy enough, but the main part of the second problem is that this segmentation part is all python/numpy/scipy, and the people in my lab I could recruit to help in this don't know python. I could teach them, but is there a better/easier way?
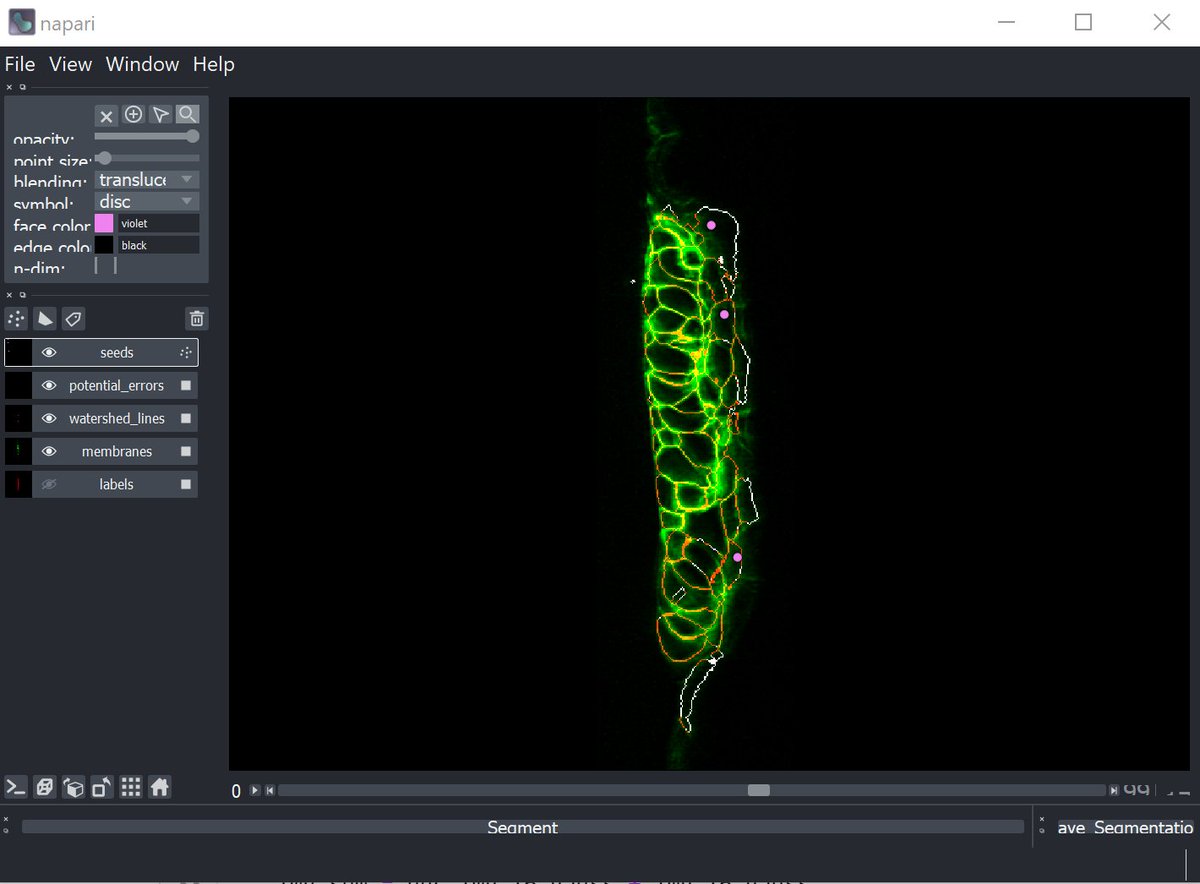
This is where Napari comes in. With it, we can easily build a GUI on top of this that lets the people in my lab who aren't comfortable with code get going right away.
We generate a napari viewer, importing the original image (green), the segmentation (red), the seed points (purple) and a calculated segmentation "confidence" image (white). (Apologies to @Red_Green_Cow)
You can see the cells that shouldn't have been segmented (with strong signals in the white confidence channel), and also their seed positions, pulled from the numpy seed array.
From here it's just a simple matter of selecting the seeds and deleting the incorrect ones. MagicGUI means we can easily create a button to pull down all the new seeds and use those to re-run the segmentation, displaying the new image.
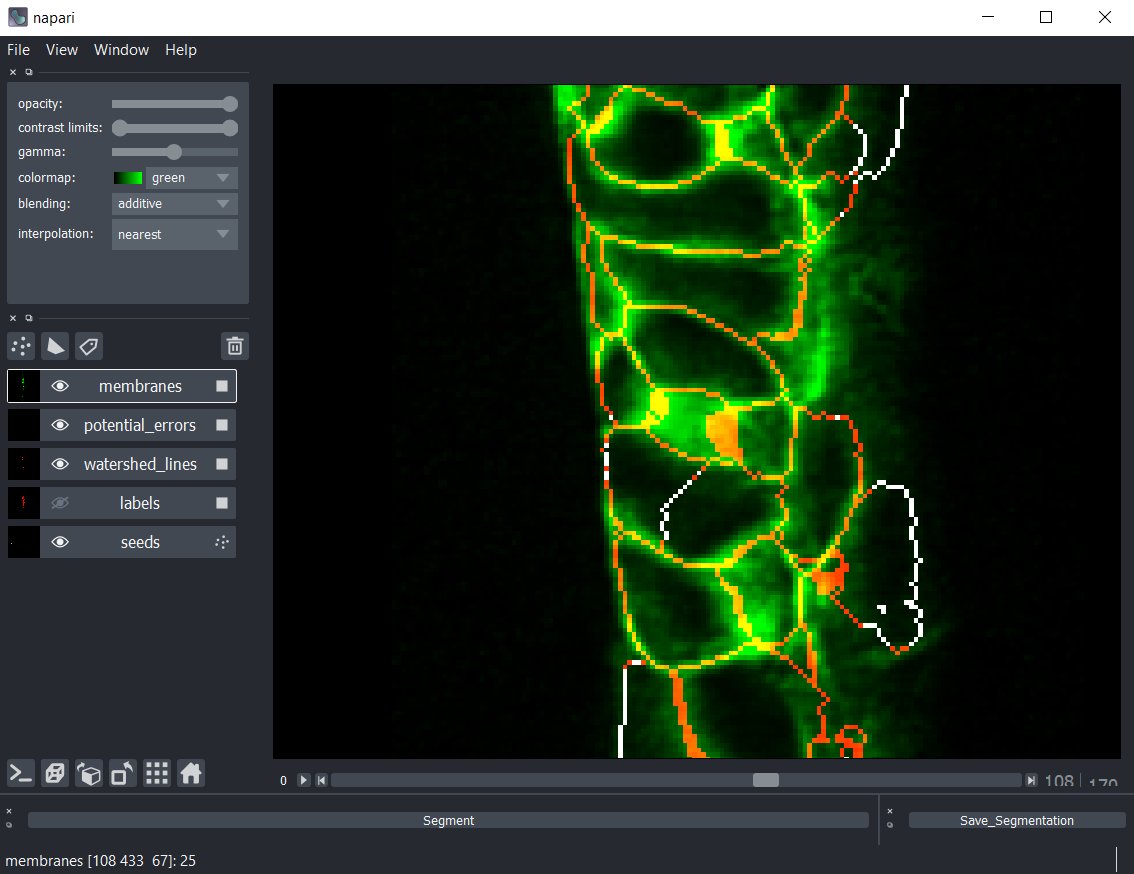
Even difficult errors, like this one near the middle, are easy to see in the viewer. Here a cell has erroneously been assigned two seed points due to a very amorphous nucleus.
It's easier to see if we use Napari to just turn on and off the confidence track -- there's no membrane there, this is just a watershed artifact from having two seeds in the same cell. We can, just like before, remove the bad seed point, and resegment and fix the error.
We can have the interns go through each image and fix the segmentation errors without touching a line of python code, just using the napari viewer, but have everything still in our python stack.
I love these kinds of enabling technologies -- the ones that not only make us work better and faster as bioimage analysts, but the ones that let us iterate quickly and easily push out the tools we've made to other people who could benefit from them.

 Read on Twitter
Read on Twitter