What I don't understand is that she claims that the #covid virus has been patented. So we cannot patent a virus if it is #natural, that would be nonsense. So the logic to have a #patent on a #virus it must be #manufactured? https://twitter.com/Smackenziekerr/status/1325804404713291779
#SARSCoV2 is a new human #coronavirus ( #CoV), which emerged in China in late 2019 and is responsible for the global #COVID19 pandemic that caused more than 540,000 deaths in six months.
Understanding the origin of this virus is an important issue and it is necessary to determine the mechanisms of its dissemination in order to contain future epidemics.
Based on phylogenetic inferences, sequence analysis and structure-function relationships of coronavirus proteins, informed by the knowledge currently available on the virus, we discuss the different scenarios evoked to account for the origin - natural or synthetic - of the virus.
Based on available data, it is currently impossible to firmly assert whether SARS-CoV2 results from a natural zoonotic emergence or from an accidental escape of a laboratory strain.
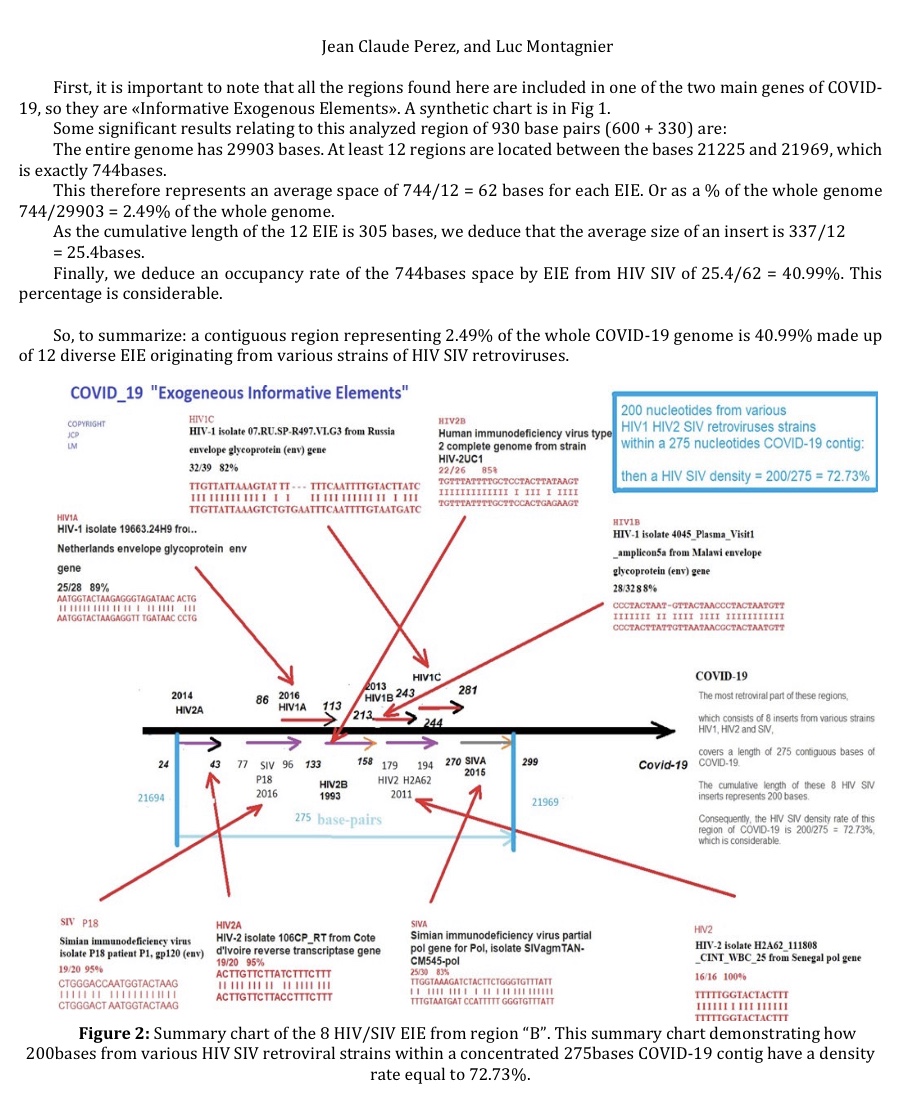
Regardless of its origin, studying the evolution of the molecular mechanisms involved in the emergence of this pandemic virus is essential to develop therapeutic and vaccine strategies.
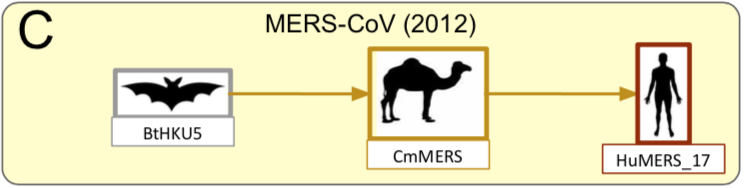
SARS-CoV-2 is the third human coronavirus (CoV) responsible for severe respiratory syndrome that emerged in the last 20 years, the two previous ones being SARS-CoV in 2002 (Drosten et al. 2003) and MERS-CoV in 2012 (Zaki et al. 2012).
SARS-CoV-2, which causes the COVID-19 disease in humans, spread into a pandemic in early 2020. By July 5, 2020, more than 11.2 million infections had been reported with at least 530,000 deaths.
The etiological agent of COVID-19 was rapidly identified and by 26 January 2020, 10 viral genomes had been sequenced (R. Lu et al. 2020). Sequence comparisons revealed a 99.98% pairwise identity between those genomes, which is characteristic of a recent emergence.
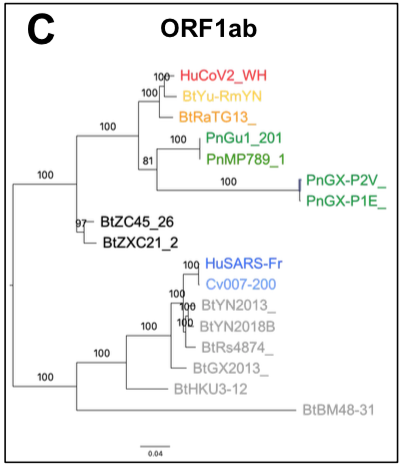
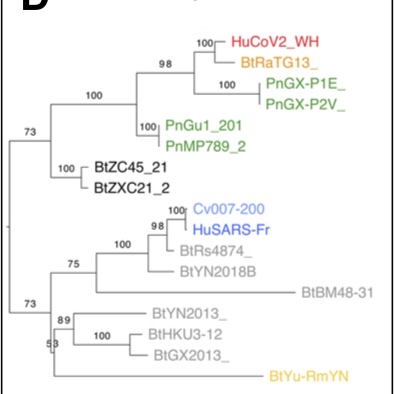
When the first SARS-CoV-2 isolates were sequenced, the closest coronaviruses available in databases were bat-SL-CoVZXC21 and bat-SL-CoVZC45 strains,
isolated in 2015 and 2017 from bats in the Zhoushan region of eastern China, and whose genomes showed 88% identity with SARS-CoV-2 (R. Lu et al. 2020).
The SARS-CoV-2 genome sequence is more distant from SARS-CoV (79% identity) and MERS-CoV (50% identity), the viruses responsible for the previous human epidemics.
Researchers concluded that SARS-CoV-2 is a new infectious agent belonging to the SARS-CoV family, able of human-to-human transmission, and whose animal reservoir is the bat (P. Zhou et al. 2020; R. Lu et al. 2020).
The zoonotic origin of CoVs is well documented. This family of viruses infects more than 500 species of chiropterans (a mammalian order consisting of more than 1200 species of bats) which represent an important reservoir for CoV evolution,
allowing the recombination of viral genomes in animals co-infected by different strains (Menachery et al. 2015; Hu et al. 2017; Luk et al. 2019).
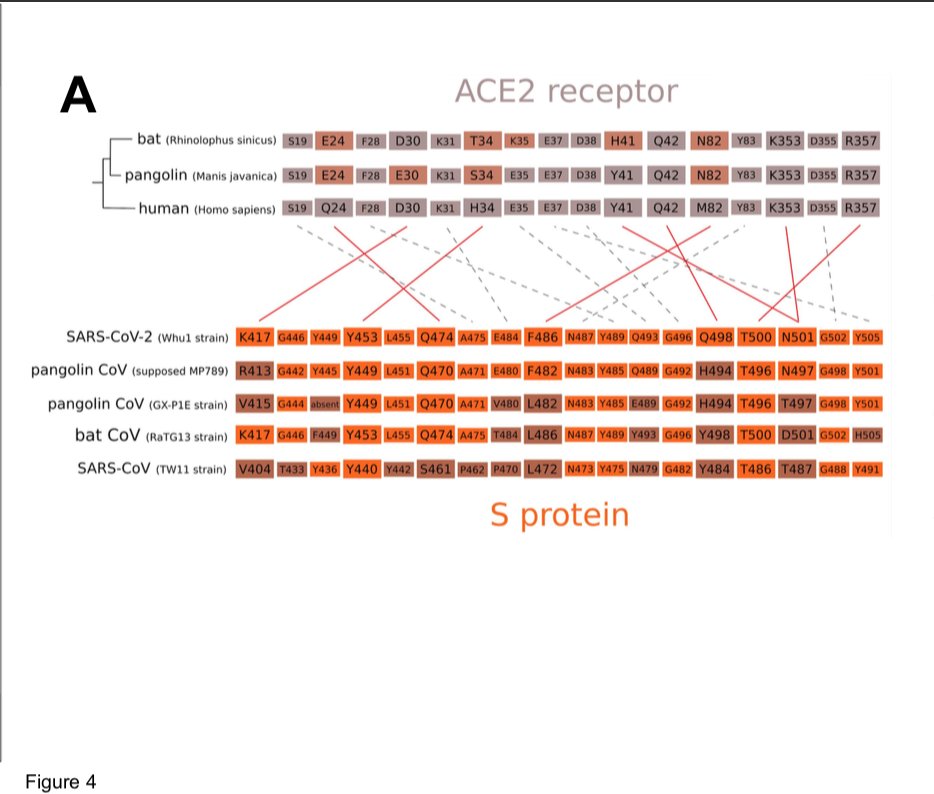
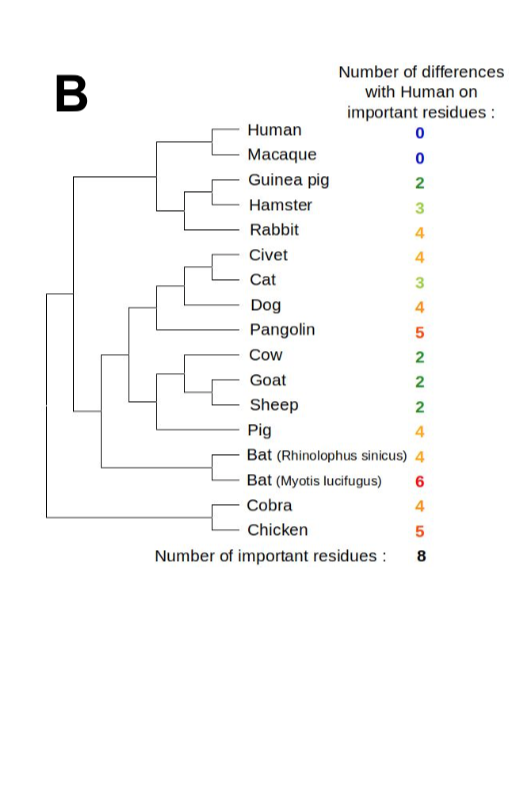
It is generally accepted that zoonotic transmission of CoVs to humans occurs through an intermediate host species, in which viruses better adapted to human receptors can be selected, thereby facilitating the species barrier crossing (Cui, Li, et Shi 2019).
Vectors of zoonotic transmission can be identified by examining the phylogenetic relationships between new viruses and viruses isolated from animal species living in the regions of emergence.
Although no epidemic related to direct bat-to-human transmission has been identified to date, experimental studies have shown that more than 60 chiropteran CoVs are capable of infecting cultured human cells (Luis et al. 2013; Menachery et al. 2015).
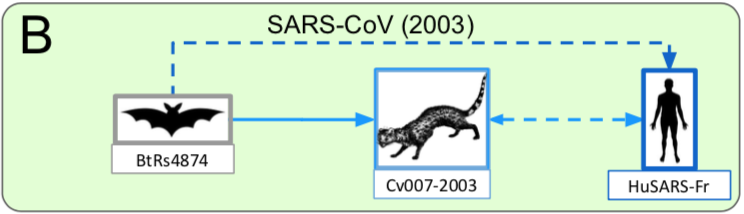
The identification, in 2017, of viral isolates very similar to SARS-CoV in bats raises the issue of a possible direct transmission from chiropterans to humans,
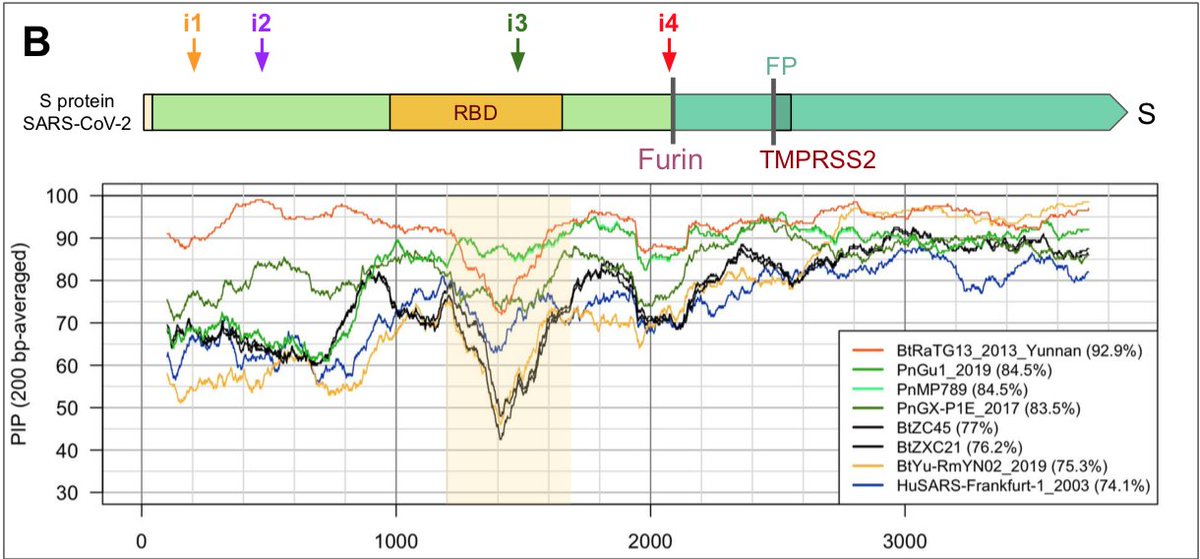
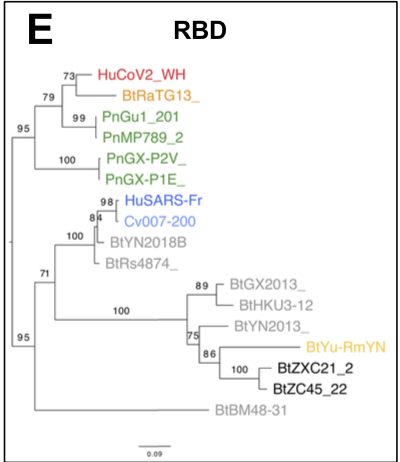
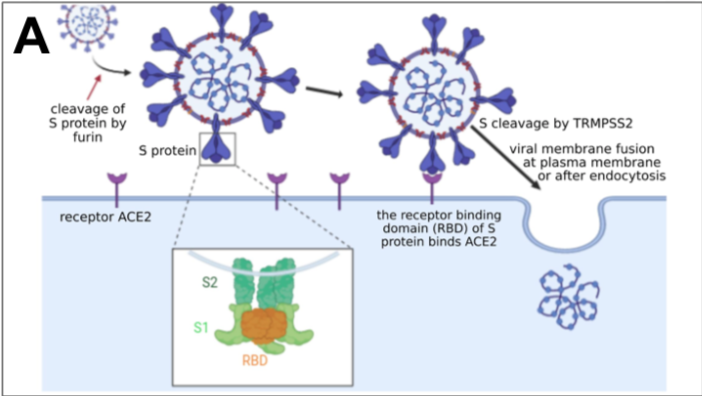
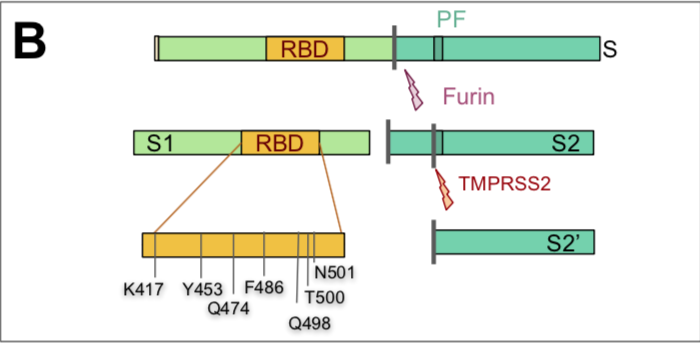
which could result from mutations in the receptor-binding domain (RBD) of the viral spike protein, which enables its entry into the host cell (Hu et al. 2017).
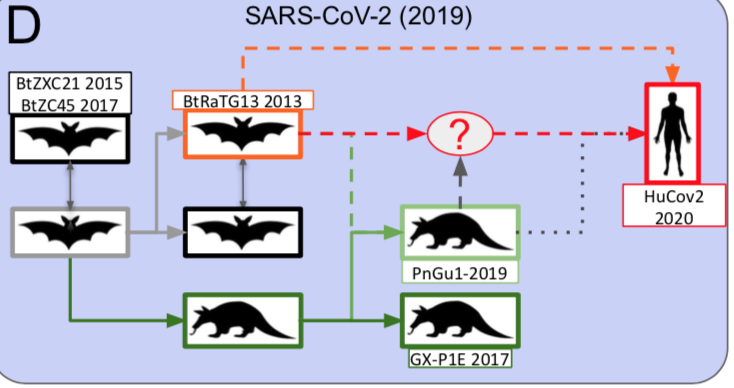
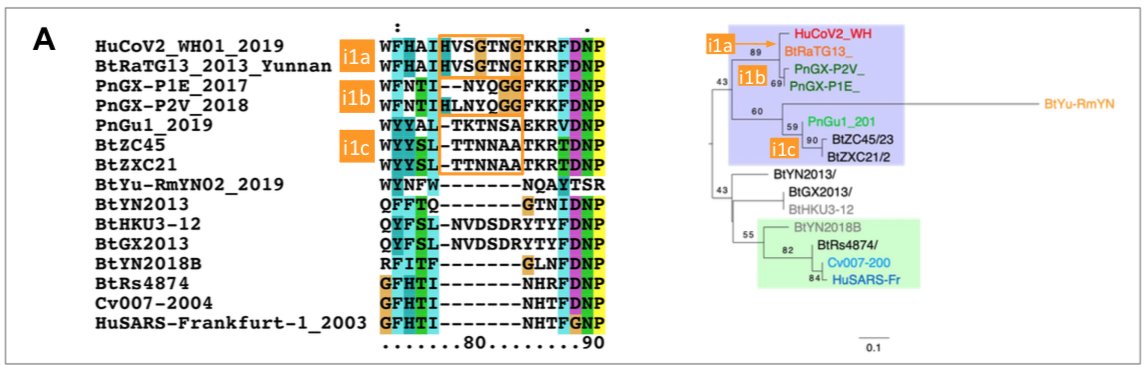
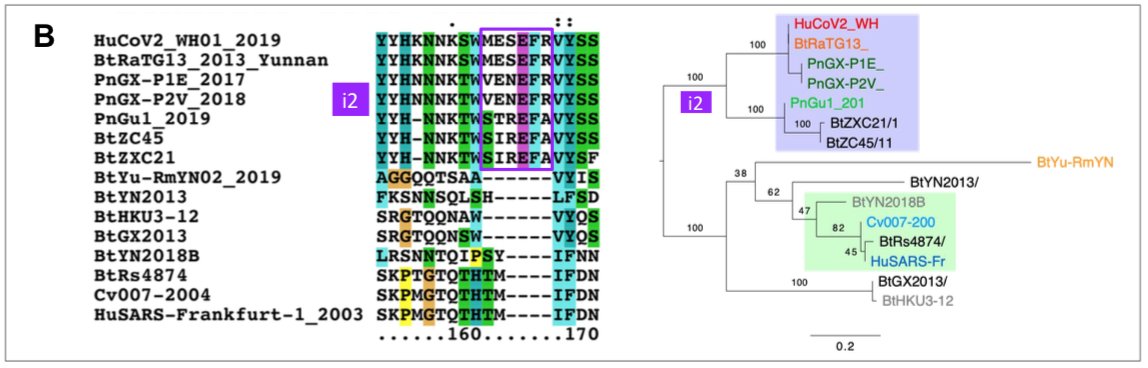
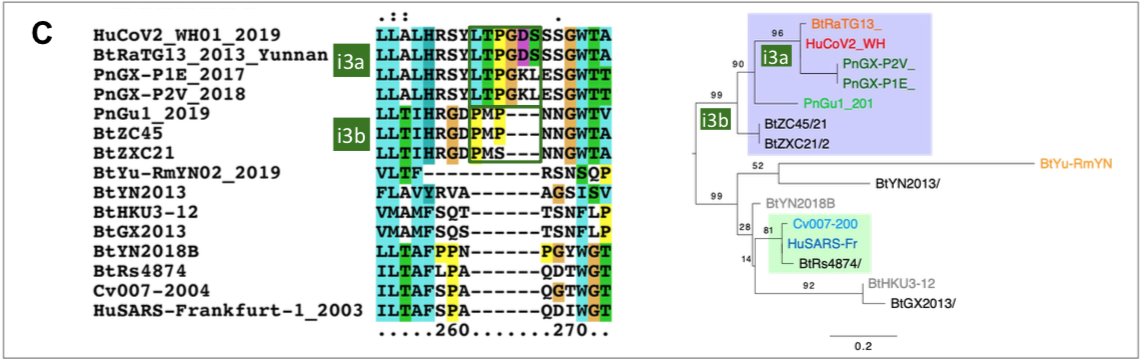
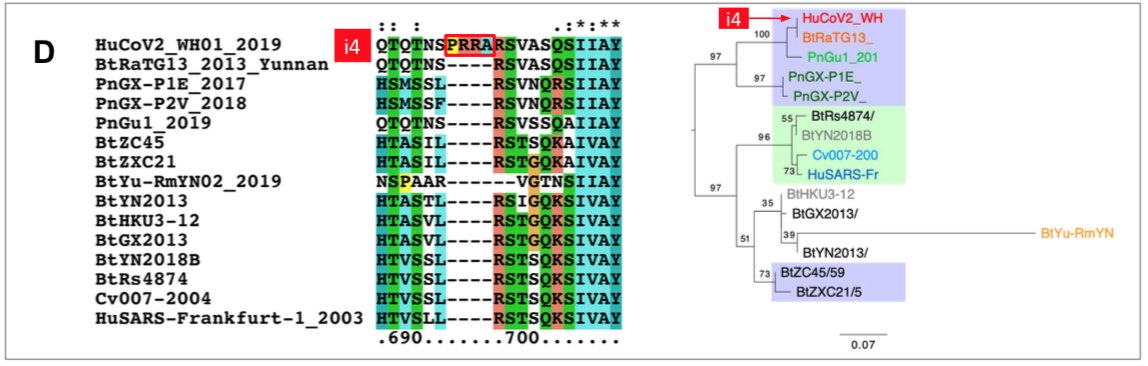
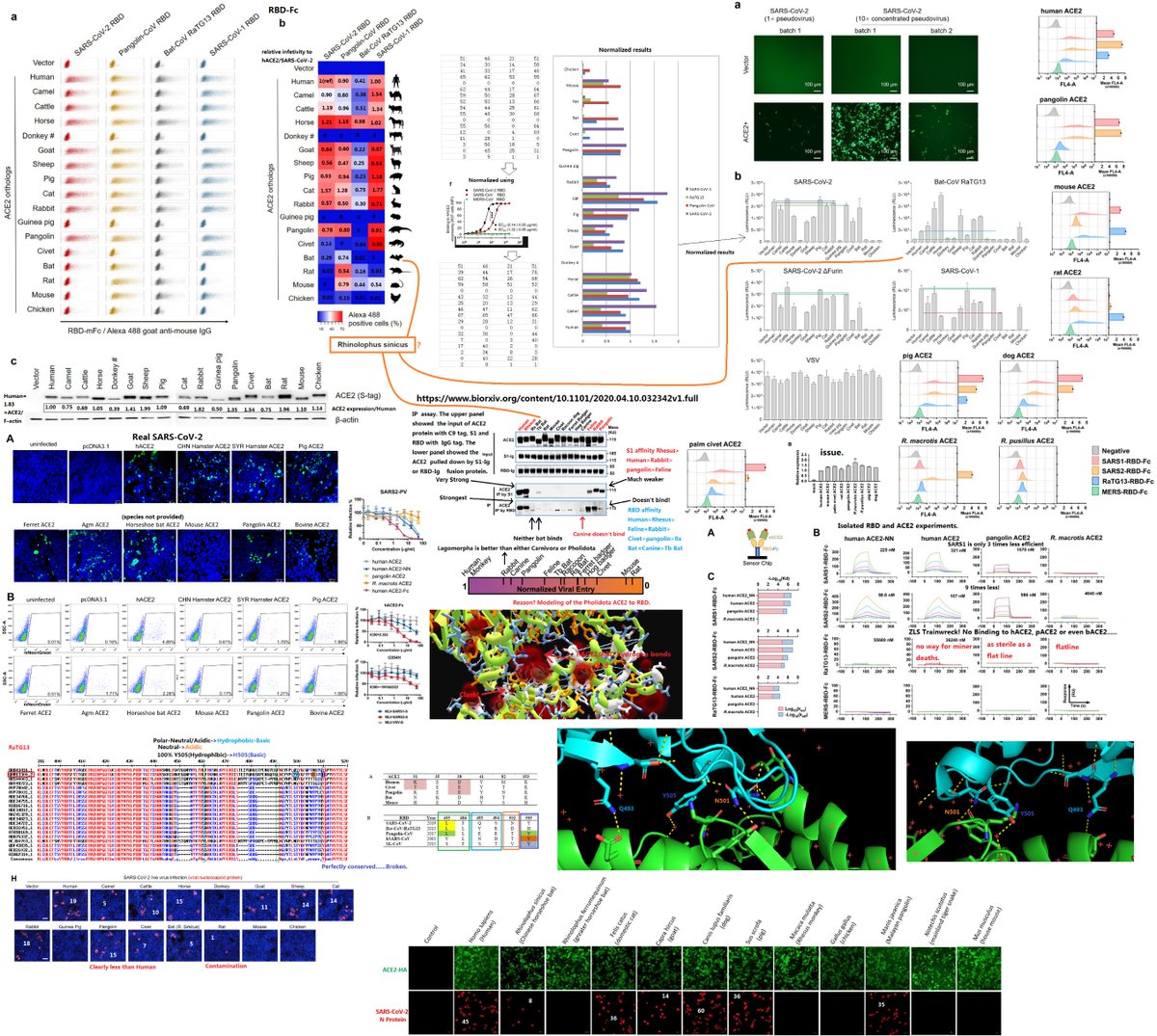
The origin of SARS-CoV-2 is a matter of debate. Bioinformatic studies revealed that it has a 96.2% identity with a CoV genome (RaTG13) reconstructed from faeces and anal samples of Rhinolophus affinis bats).
Interestingly, these samples were collected in 2013, but the full genome sequence was only published in early February 2020 (P. Zhou et al. 2020). Unfortunately, the precise location of the sample collection is documented neither in the article nor in the sequence databases.
However, we found an exact match between RaTG13 and a 370 nucleotide fragment published in 2016 (KP876546), encoding a BtCoV/4991 polymerase domain,
which had been sequenced from isolates collected from a mine shaft in Yunnan Province following the death of 3 miners from an atypical pneumonia (Ge et al. 2016).
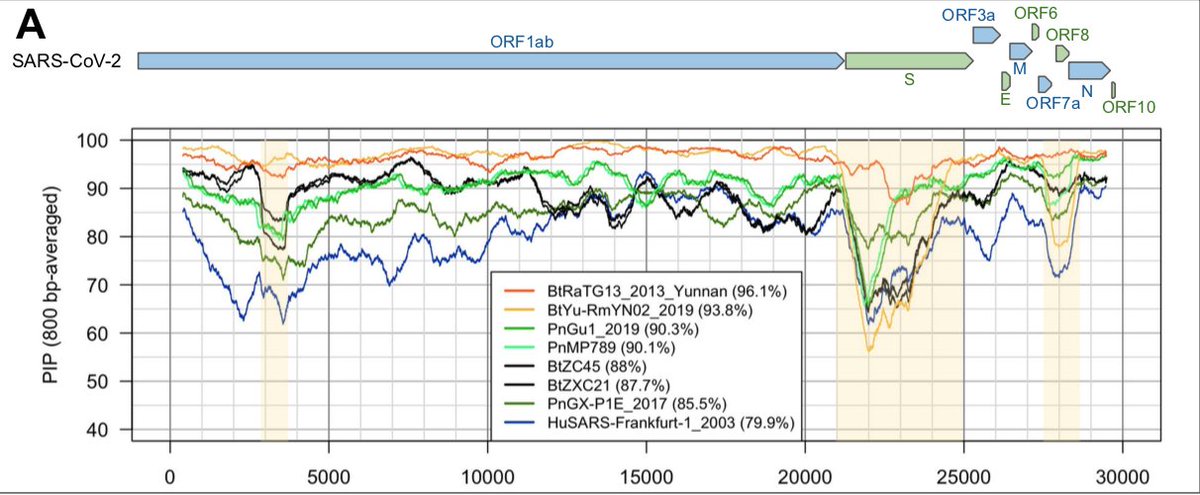
More recently, a metagenome (RmYN02) was assembled from faeces samples of 11 bats of the species Rhinolophus malayanus, collected in 2019 in Yunnan province. This sequence has 97.2% identity with the first two thirds of the SARS-CoV-2 (ORF 1ab) genome.
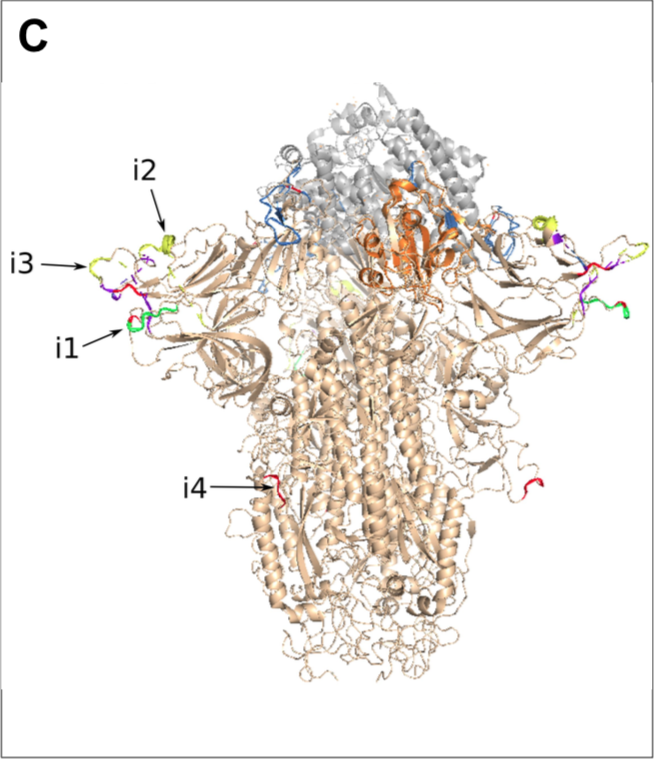
However, on the remaining third of the genome it diverges quite strongly, especially at the level of the S1 protein and ORF 8 (Figure 2) (H. Zhou et al. 2020).
The length of CoV genomes is about 30,000 nucleotides, which is exceptionally long for an RNA virus (by comparison, the length of AIDS and Ebola virus genomes are about 10,000 and 19,000 nucleotides, respectively).
CoVs are able to maintain such a long a genome thanks to a replication error correction system unique in the world of RNA viruses,
depending on a viral exonuclease that ensured a proofreading mechanism limiting the mutation rate (Eckerle et al. 2010; Ferron et al. 2018; Casane, Policarpo, et Laurenti 2019).
The first two-thirds of the genome corresponds to a single gene, ORF1ab, coding for a polyprotein precursor, which is then cleaved into 16 proteins forming the replication/transcription complex. https://zenodo.org/record/3724003#.X6oBIy1h1QI
The last third contains 9 genes coding for proteins produced from subgenomic RNAs synthesized by viral polymerase (Figure 2A).
@threader_app compile

 Read on Twitter
Read on Twitter