My first first author publication got published at @NatureComms! 

Together with @DruSalva @ucielp , Juli Mateos, @lamozamalaonda @NiGB26 Arnaldo, Edgardo, @FitoDeRosario @dmoreno26 and @JavierPalatnik
Thread https://twitter.com/NatureComms/status/1319661105254264832
https://twitter.com/NatureComms/status/1319661105254264832


Together with @DruSalva @ucielp , Juli Mateos, @lamozamalaonda @NiGB26 Arnaldo, Edgardo, @FitoDeRosario @dmoreno26 and @JavierPalatnik
Thread
 https://twitter.com/NatureComms/status/1319661105254264832
https://twitter.com/NatureComms/status/1319661105254264832
In the @rnabiollab at @IBR_CONICET we study how plant MIRNA are processed


http://www.ibr-conicet.gov.ar/en/laboratorios/palatnik


http://www.ibr-conicet.gov.ar/en/laboratorios/palatnik
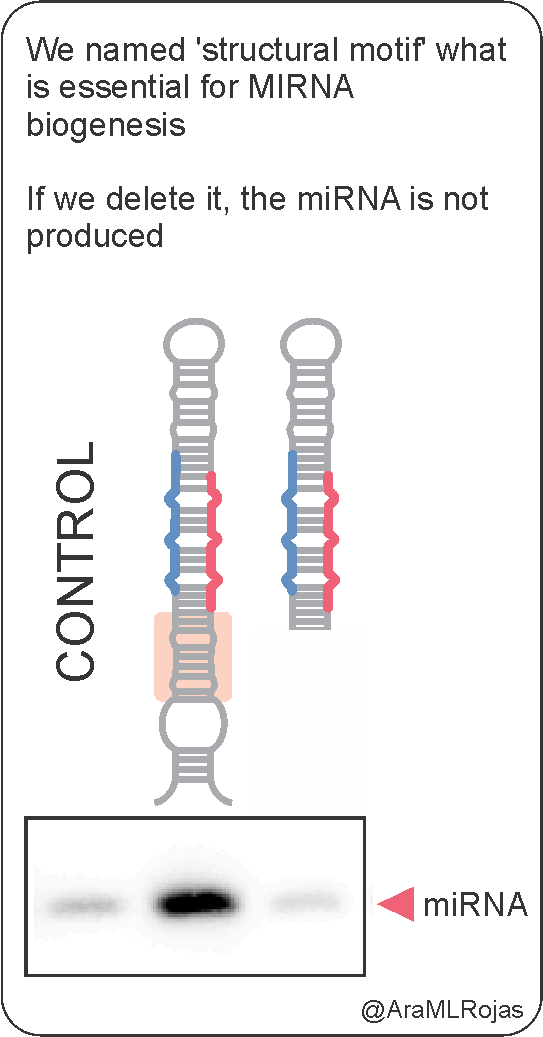
In previous works from our Lab as well as other groups, it was described the diversity of plant MIRNA precursors and the structural motifs that are necessary for processing and microRNA release
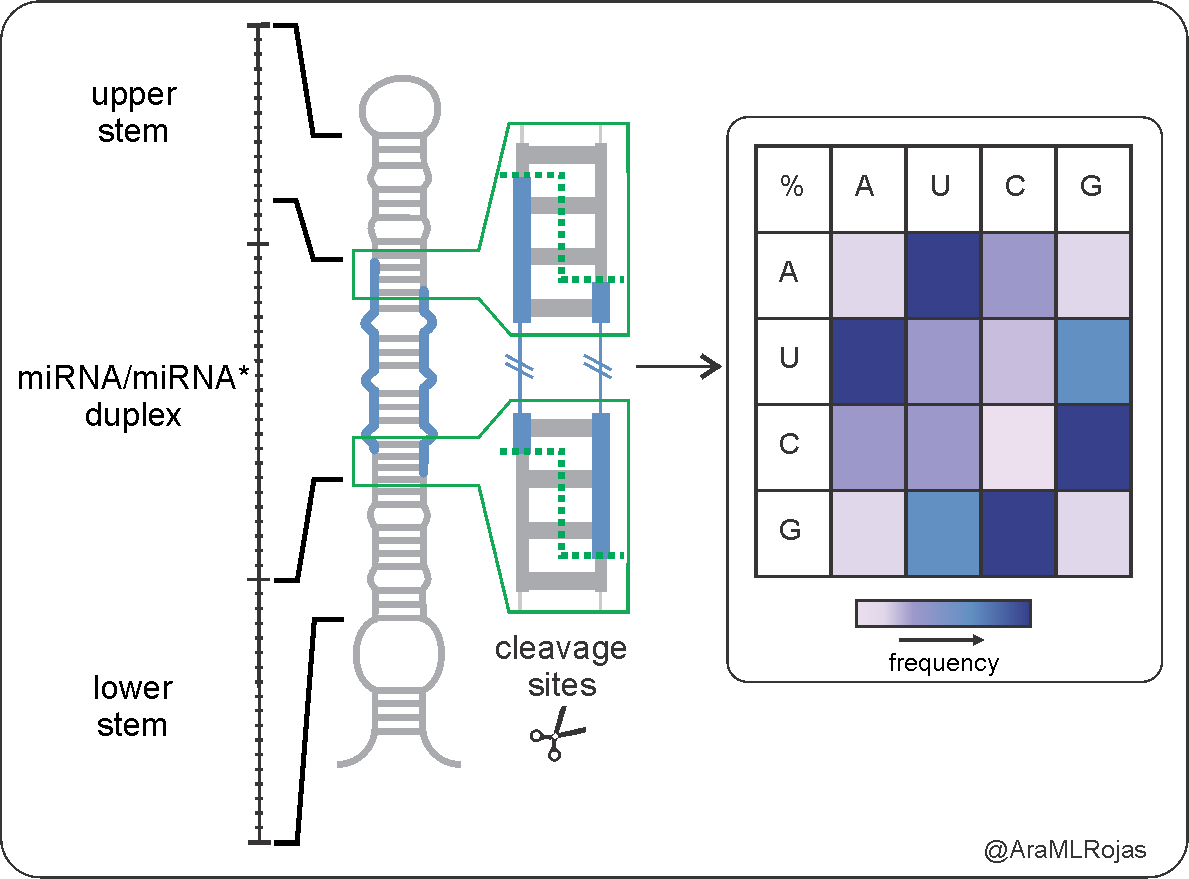
In our latest paper we could show that the identity of the nucleotides at Dicer cleavage sites are important for processing efficiency https://www.nature.com/articles/s41467-020-19129-6
During my PhD, I realized that what I love the most of any RNA molecule is all the information that can store. For me was amazing to find out, after a lot of aligning and digging, that plant MIRNA precursors present biases at the DCL1 cleavage sites 



These biases are important because they affect the efficiency of MIRNA processing, which is reflected in the amount of microRNA produced from a precursor.
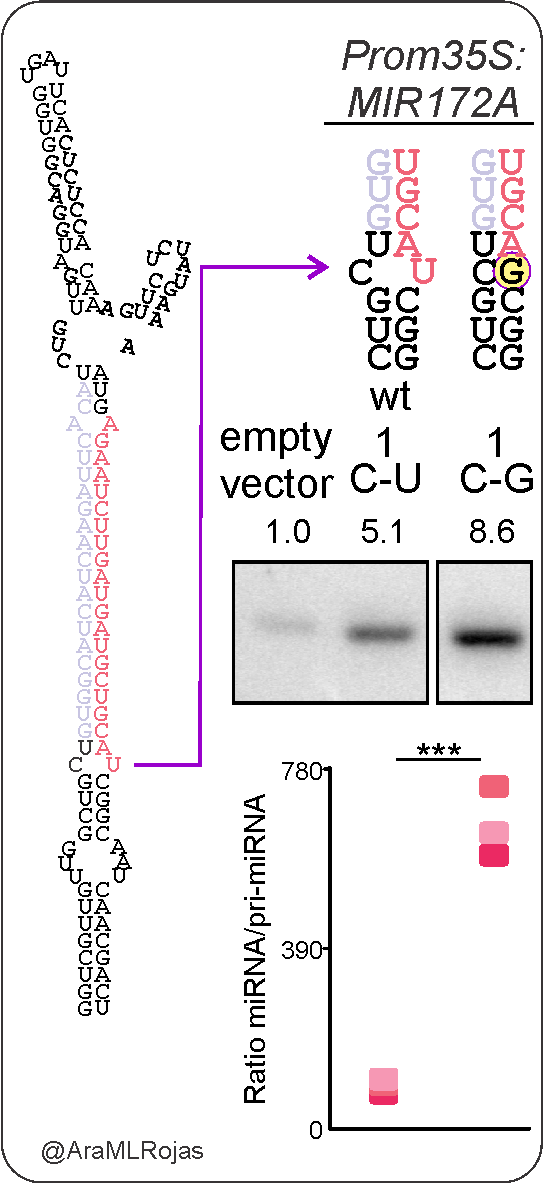
With a single nucleotide change, from C-U to C-G in the first cleavage site, we could enhance MIR172A processing.
With a single nucleotide change, from C-U to C-G in the first cleavage site, we could enhance MIR172A processing.
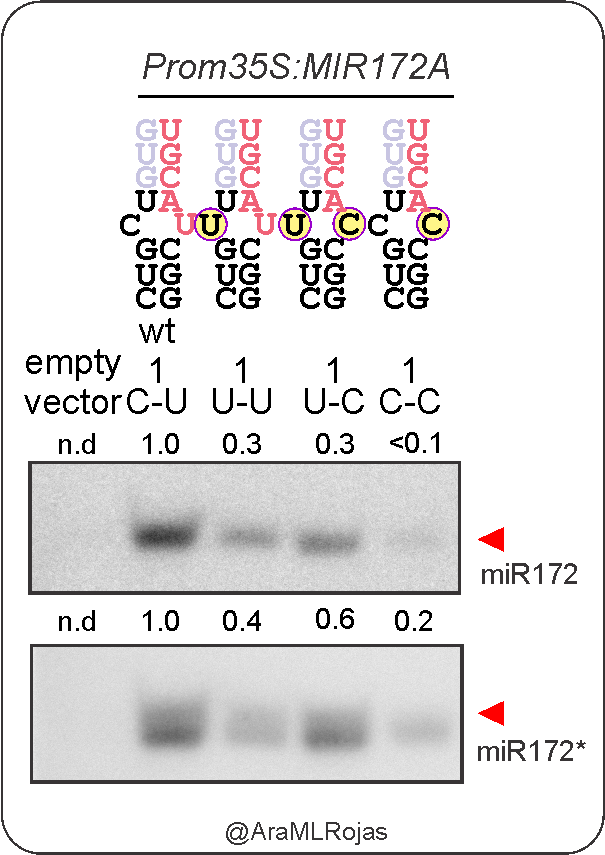
The identity of nucleotides at mismatched positions plays a crucial role on processing efficiency as well.
C-C mismatches are “malo, malo, muy malo” for processing, and spoiler alert, they can only be found at low frequency in plant MIRNA precursors and other structured RNAs
C-C mismatches are “malo, malo, muy malo” for processing, and spoiler alert, they can only be found at low frequency in plant MIRNA precursors and other structured RNAs
To understand why, in collaboration with @dmoreno26 and @DruSalva we studied the behavior of different nucleotide pairs in MIR172A by Molecular Dynamics.
The C-U mismatch (wt) is stabilized half of the simulation time
@DruSalva
The C-U mismatch (wt) is stabilized half of the simulation time
@DruSalva
But we found that C-C mismatches really like to dance! They move a lot more when compared to other nucleotide pairs 

@DruSalva


@DruSalva
Thanks to all of those who helps us along the way, specially to the co-authors, lab mates and friends who supported me during this project!
Thanks to @marlene_reichel from @StaigerLab who invited me last year after we met at RNA2019 meeting to share this story with them.
to share this story with them.
As well as to @CommsBio and @RNASociety for the travel grants that allowed me to be there!
 to share this story with them.
to share this story with them. As well as to @CommsBio and @RNASociety for the travel grants that allowed me to be there!
Finally, thanks to the @NatureComms Editor and reviewers for the amazing reviewing process, even though the COVID situation
Since we cannot have the usual barbeque to celebrate because COVID, I had to wear the crown in the lab, and I received some drinks from @CamilaGoldy (aka my elbow sister) to enjoy at home 


 Read on Twitter
Read on Twitter