Tired of clicking the refresh button on the NYtimes election update page? Try clicking here and reading our new preprint instead: https://www.biorxiv.org/content/10.1101/2020.11.04.368217v1
This was a fun project with lots of great scientists including @ackero9, @pmberube,
@th4ckl, @rstepanauskas, Penny and Dan!
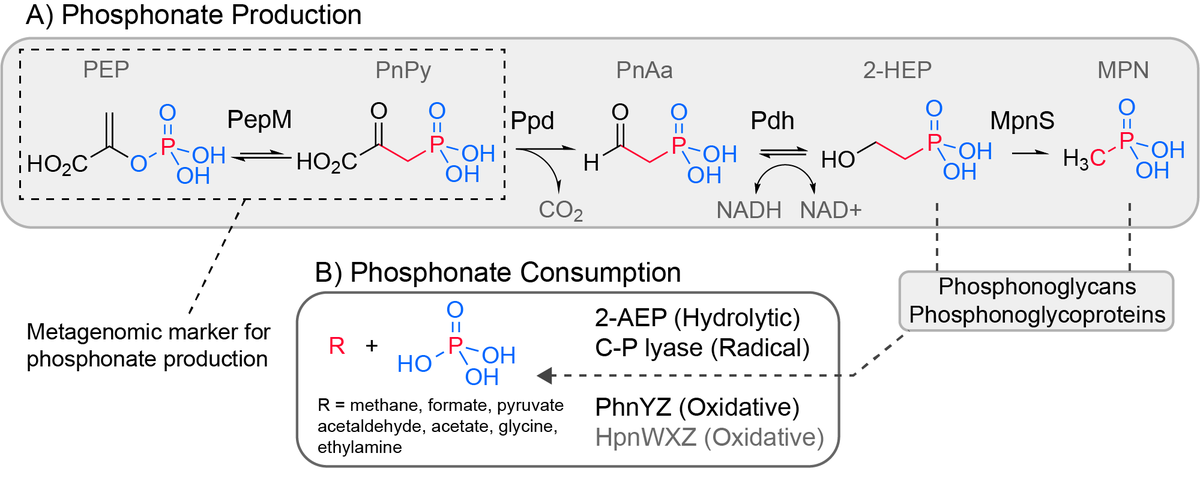
We wanted to know who makes phosphonates (reduced phosphorus compounds with a carbon phosphorus bond) in the upper 200 meters of the ocean and why.
@th4ckl, @rstepanauskas, Penny and Dan!
We wanted to know who makes phosphonates (reduced phosphorus compounds with a carbon phosphorus bond) in the upper 200 meters of the ocean and why.
phosphonates are an enigmatic and abundant (20-25% of DOP) organic phosphorus pool in the ocean. Microbial degradation of methylphosphonate can account for methane supersaturation in oxygenated upper ocean, thus linking phosphonates to greenhouse gasses.
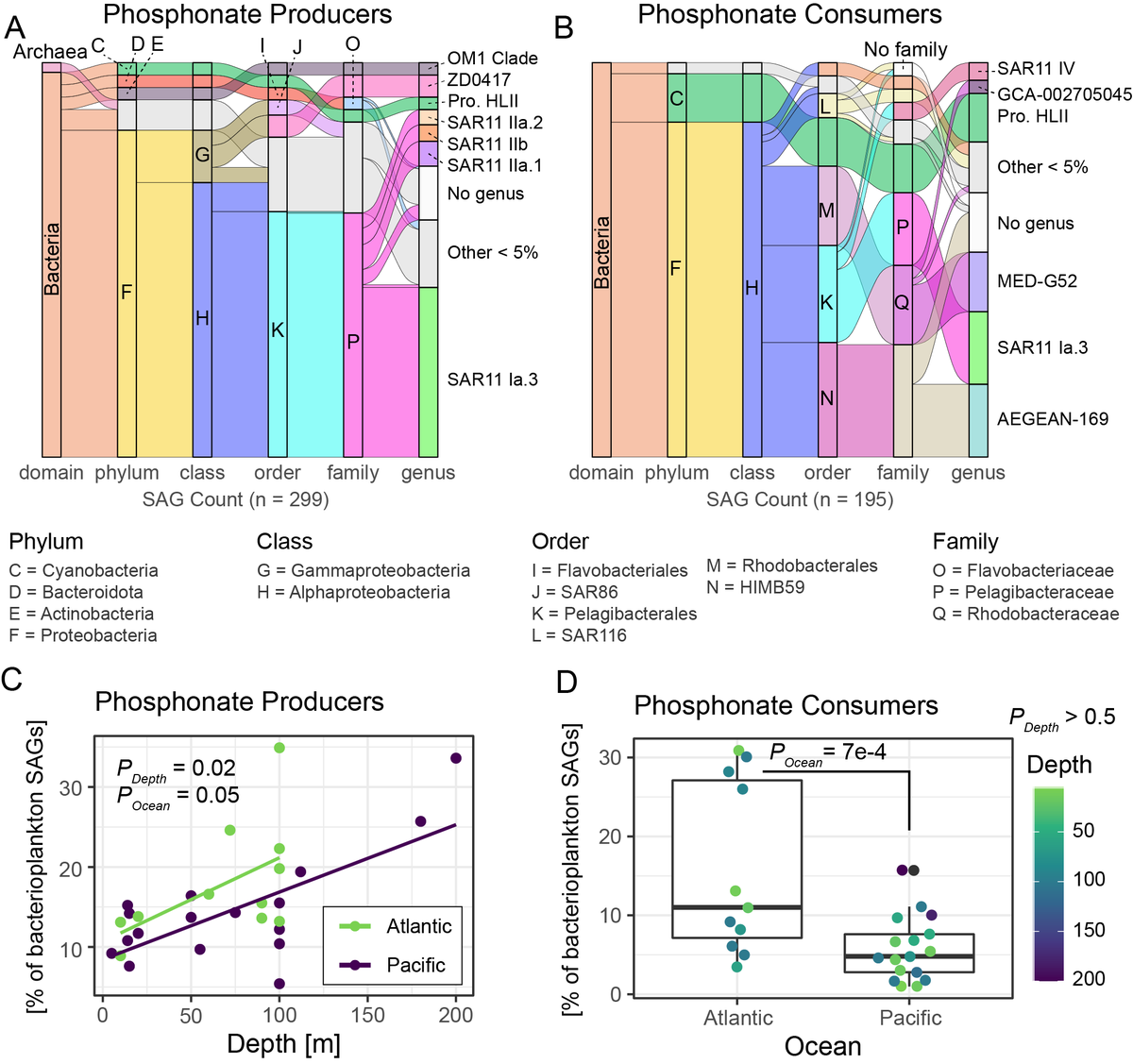
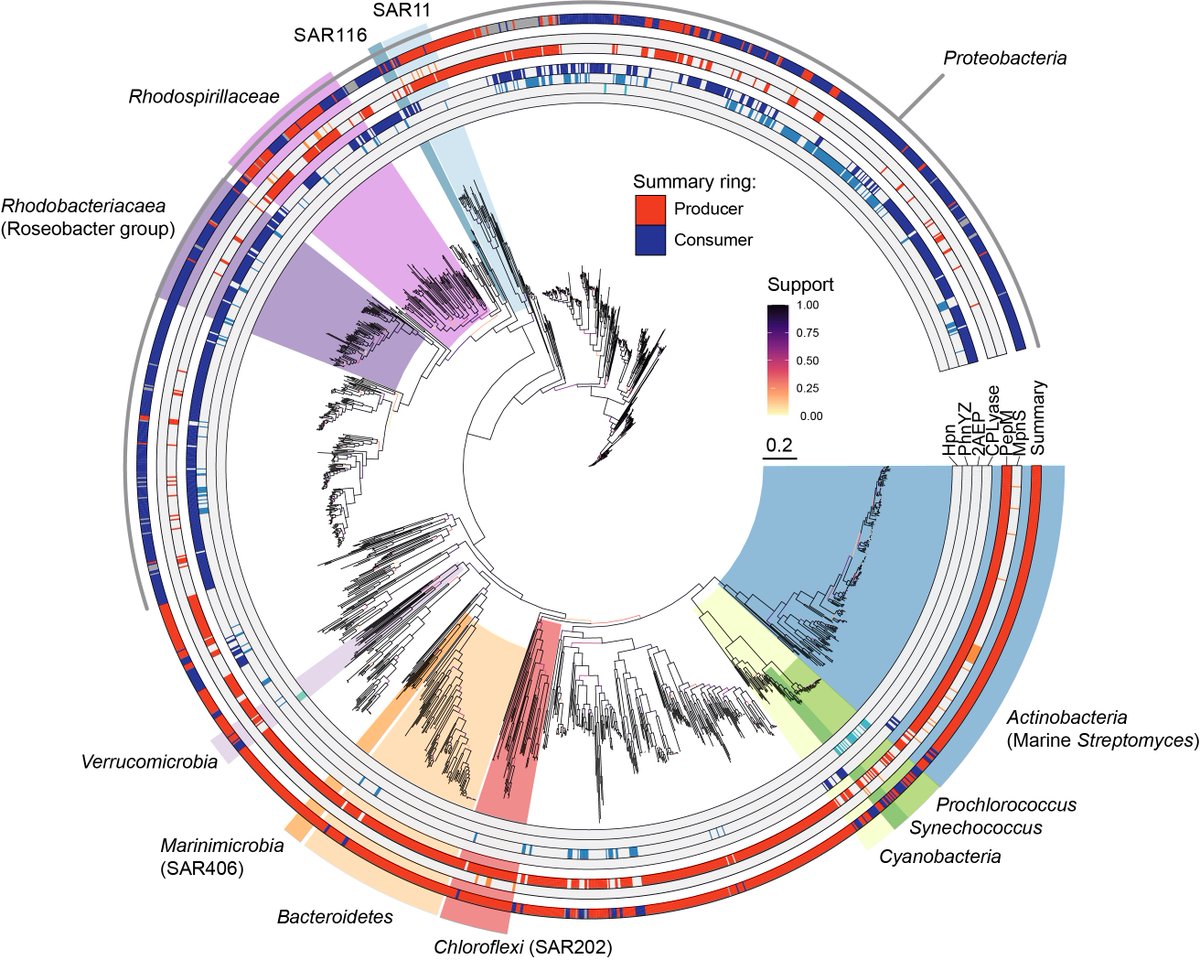
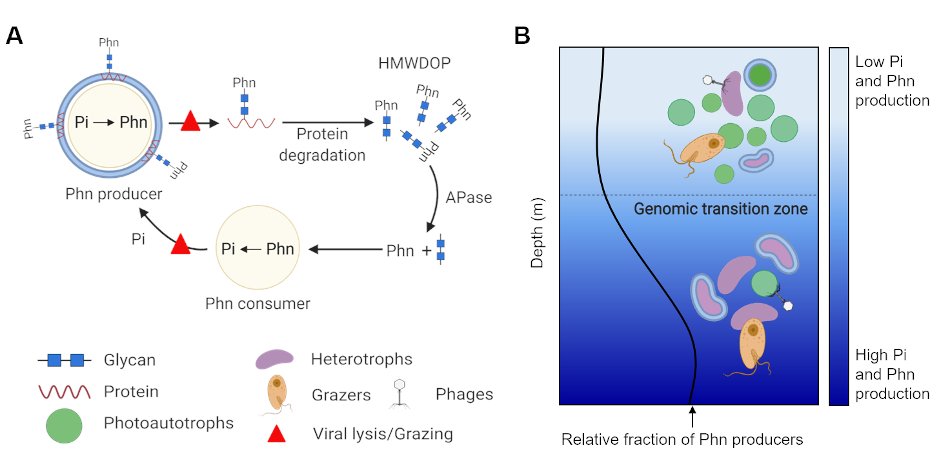
The biosynthetic enzymes are known so we could search for them in genomes. We looked into over 13000 genomes from the the GORG-tropics dataset and found that SAR11 and Prochlorococcus are abundant producers.
We also found that phosphonate producers increased with increasing depth in metagenomes - peaking near the DCM and what has been termed the "genomic transition zone."
At the time we started this project there was only one surface ocean, oligotrophic, free-living marine bacterial strain with phosphonate biosynthesis genes in culture. It happened to be a Prochlorococcus strain from our culture collection!
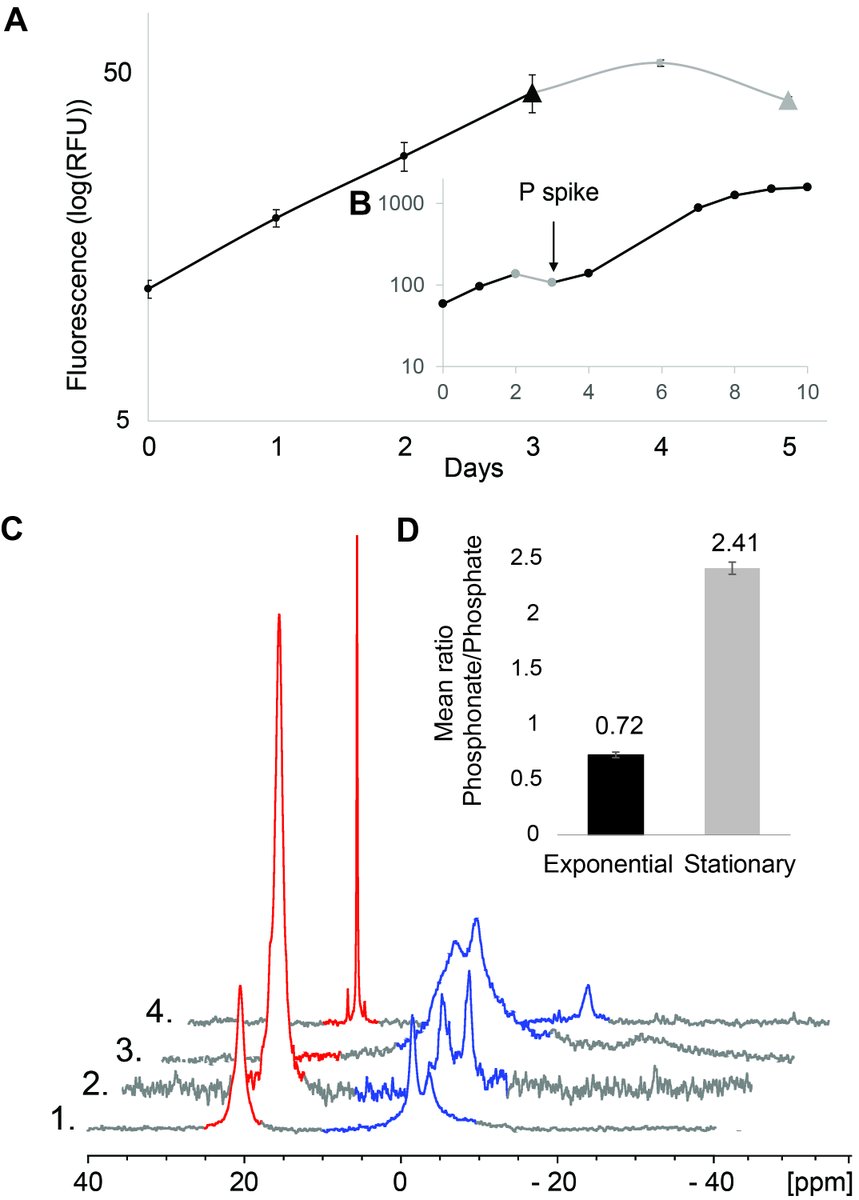
Turns out that Prochlorococcus can make A LOT of phosphonates - in some cases devoting up to 50% of cellular P quotas towards phosphonate production.
Why would Prochlorococcus put so much P into phosphonate? Could it be a luxury storage strategy? We don't think so.
Why would Prochlorococcus put so much P into phosphonate? Could it be a luxury storage strategy? We don't think so.
Prochlorococcus doesn't rapidly consume its phosphonate reservoir upon P starvation. Prochlorococcus phosphonate producers also do not appear to have the genes necessary to break C-P bonds.
In fact, effectively no marine phosphonate producing genomes have the genes required to break C-P bonds. So if you are a phosphonate producer in the ocean it is unclear how you could get your P back from phosphonates once it is locked away in these compounds.
All of Prochlorococcus' phosphonates were in the high molecular weight protein fraction. We think this is because phosphonates are incorporated into sugar chains that are attached to anchor proteins embedded in the outer membrane.
Gene context in other genomes suggests that phosphonglycoproteins are a common cell surface modification in the ocean. We think that this is probably because they are a kind of phage attachment or grazer defense mechanism.
Disguising or modifying the cell surface is a common strategy for pathogenic bacteria to avoid the immune system and in developing resistance to phage. We think a large part of the dissolved organic phosphorus in the ocean might form as a consequence of this defense strategy.
Thanks for reading and check out the preprint. We welcome feedback. I think @ackero9 has follow up studies in the works :)

 Read on Twitter
Read on Twitter