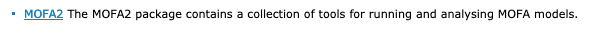Some of the new Bioconductor packages that caught my eye. There are way more great new contributions than I can cover, do check the full list here for your topics of interest:
http://bioconductor.org/news/bioc_3_12_release/ https://twitter.com/mikelove/status/1323247834871795714
http://bioconductor.org/news/bioc_3_12_release/ https://twitter.com/mikelove/status/1323247834871795714
bambu from @chenyin76951822 Yuk Kei Wan @JonathanGoeke
multi-sample transcript discovery and quantification using long read RNA-Seq data
http://bioconductor.org/packages/release/bioc/html/bambu.html
multi-sample transcript discovery and quantification using long read RNA-Seq data
http://bioconductor.org/packages/release/bioc/html/bambu.html
biocthis from @lcolladotor and @M2RUseR
expands the usethis package with the goal of helping automate the process of creating R packages for Bioconductor
http://bioconductor.org/packages/release/bioc/html/biocthis.html
expands the usethis package with the goal of helping automate the process of creating R packages for Bioconductor
http://bioconductor.org/packages/release/bioc/html/biocthis.html
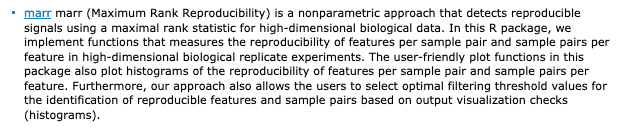
marr from Katerina Kechris and @ghoshd groups
nonparametric approach that detects reproducible signals using a maximal rank statistic for high-dimensional biological data
http://bioconductor.org/packages/release/bioc/html/marr.html
nonparametric approach that detects reproducible signals using a maximal rank statistic for high-dimensional biological data
http://bioconductor.org/packages/release/bioc/html/marr.html
MOFA2, sequel to MOFA, from @RArgelaguet @BrittaVelten and Damien Arnol, Danila Bredikhin
http://bioconductor.org/packages/release/bioc/html/MOFA2.html
http://bioconductor.org/packages/release/bioc/html/MOFA2.html
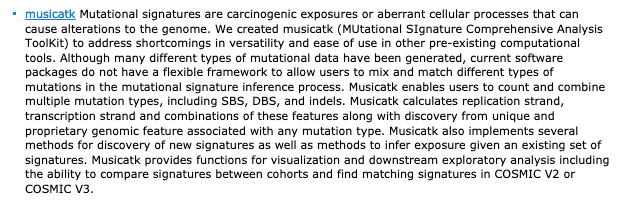
musicatk from Chevalier and Campbell
flexible framework to allow users to mix and match different types of mutations in the mutational signature inference process
http://bioconductor.org/packages/release/bioc/html/musicatk.html
flexible framework to allow users to mix and match different types of mutations in the mutational signature inference process
http://bioconductor.org/packages/release/bioc/html/musicatk.html
pipeComp from @GermainPl @AnthonySonrel @markrobinsonca
facilitate the comparison of pipelines involving various steps and parameters
http://bioconductor.org/packages/release/bioc/html/pipeComp.html
facilitate the comparison of pipelines involving various steps and parameters
http://bioconductor.org/packages/release/bioc/html/pipeComp.html
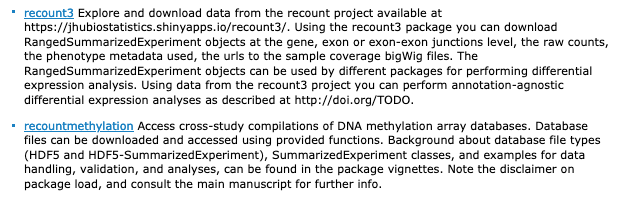
recount3 from @lcolladotor
recountmethylation from Maden, Thompson, @KasperDHansen @AbhiNellore
great compilation datasets, ready to analyze in Bioc
http://bioconductor.org/packages/release/bioc/html/recount3.html
http://bioconductor.org/packages/release/bioc/html/recountmethylation.html
recountmethylation from Maden, Thompson, @KasperDHansen @AbhiNellore
great compilation datasets, ready to analyze in Bioc
http://bioconductor.org/packages/release/bioc/html/recount3.html
http://bioconductor.org/packages/release/bioc/html/recountmethylation.html
SpatialExperiment from @drighelli @drisso1893
Bioc S4 classes for storing data for spatial experiments
http://bioconductor.org/packages/release/bioc/html/SpatialExperiment.html
Bioc S4 classes for storing data for spatial experiments
http://bioconductor.org/packages/release/bioc/html/SpatialExperiment.html
velociraptor  (i know that's a T-Rex...) from @KevinRUE67 Aaron Lun @CSoneson
(i know that's a T-Rex...) from @KevinRUE67 Aaron Lun @CSoneson
Bioconductor-friendly wrappers for RNA velocity calculations in single-cell RNA-seq data
http://bioconductor.org/packages/release/bioc/html/velociraptor.html
 (i know that's a T-Rex...) from @KevinRUE67 Aaron Lun @CSoneson
(i know that's a T-Rex...) from @KevinRUE67 Aaron Lun @CSoneson Bioconductor-friendly wrappers for RNA velocity calculations in single-cell RNA-seq data
http://bioconductor.org/packages/release/bioc/html/velociraptor.html

 Read on Twitter
Read on Twitter