 How does chemotherapy affect healthy cells in the blood? Our preprint “The evolution of hematopoietic cells under cancer therapy” just out in biorXiv! https://bit.ly/34DHplQ From @bbglab & @hematosantpau
How does chemotherapy affect healthy cells in the blood? Our preprint “The evolution of hematopoietic cells under cancer therapy” just out in biorXiv! https://bit.ly/34DHplQ From @bbglab & @hematosantpau  @a_cortesbu @fmuinos Marta Abel @nlbigas & yours truly. Tweetorial
@a_cortesbu @fmuinos Marta Abel @nlbigas & yours truly. Tweetorial
We and others have described the mutational footprint of some chemotherapies, such as cisplatin and 5-FU in metastatic cells ( https://go.nature.com/3mDcxZ8 ). We can observe it because the treatment causes mutations, and because in the metastasis a clonal expansion has occurred 

But what might be the effect of such therapies in healthy cells? Do they leave a footprint?? Do they alter the evolution of the healthy tissue??? Can we use this footprint to explore the evolution of somatic tissues????We decided to find out!
We focused on hematopoiesis. Why? We know of two conditions that have some relation to chemo exposure and involve a clonal expansion: myeloid leukemias developed after the treatment of a solid tumor (tAML), and clonal hematopoiesis (CH).
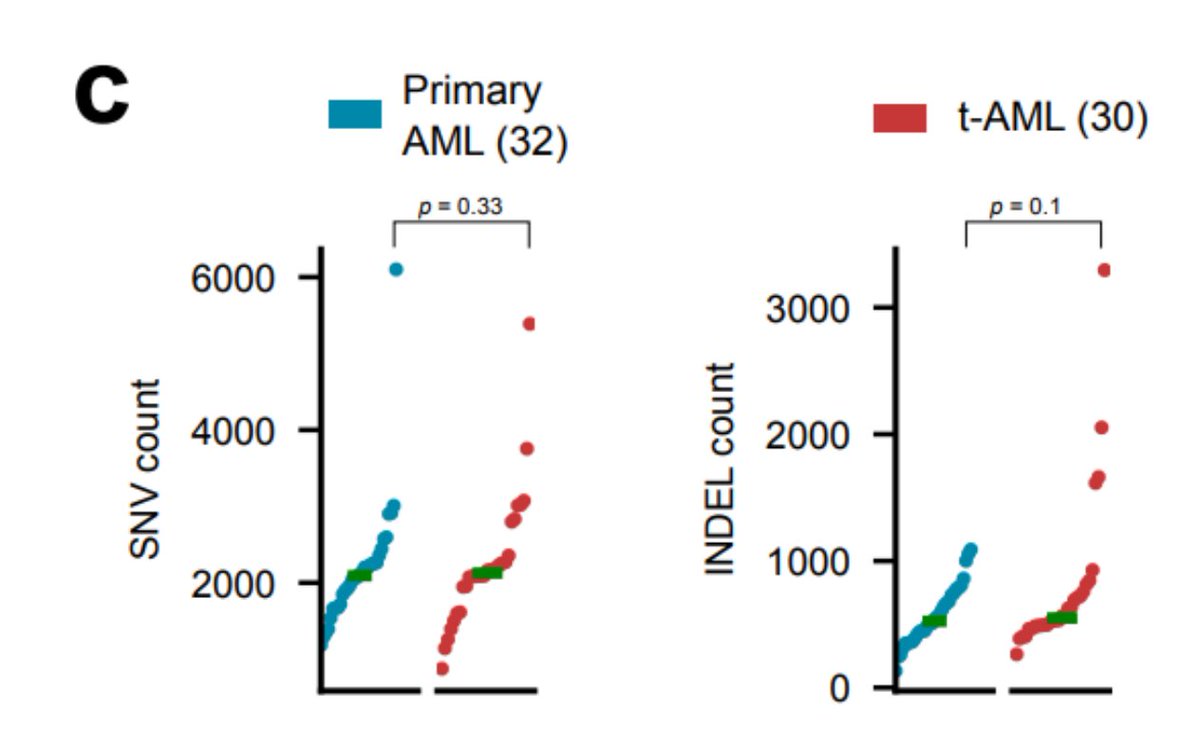
So, we obtained tAML samples @hematosantpau @a_cortesbu & some other tAML and primary AML from published studies (32 primary and 30 tAML) & identified their mutations using a uniform pipeline. As reported before, no difference in the mutation burden of both groups is seen...

But hold on!! Because we can clearly identify the mutational footprint of platinum-based drugs in tAML samples from all patients previously exposed to platinum-based cytotoxic agents!!
Because we can clearly identify the mutational footprint of platinum-based drugs in tAML samples from all patients previously exposed to platinum-based cytotoxic agents!! 

 Because we can clearly identify the mutational footprint of platinum-based drugs in tAML samples from all patients previously exposed to platinum-based cytotoxic agents!!
Because we can clearly identify the mutational footprint of platinum-based drugs in tAML samples from all patients previously exposed to platinum-based cytotoxic agents!! 

The number of platinum-related mutations is in the same order of magnitude as observed in metastatic samples, which suggests that exposure to platinum-based drugs is a sufficient condition for the acquisition of platinum-related mutations 

What about the 5-FU mutational footprint in the (3) samples exposed to the drug? We found no trace! Is it possible that hematopoietic cells do not incorporate 5-FU mutations because they are quiescent? We went back to the treated metastatic samples to try to answer this question
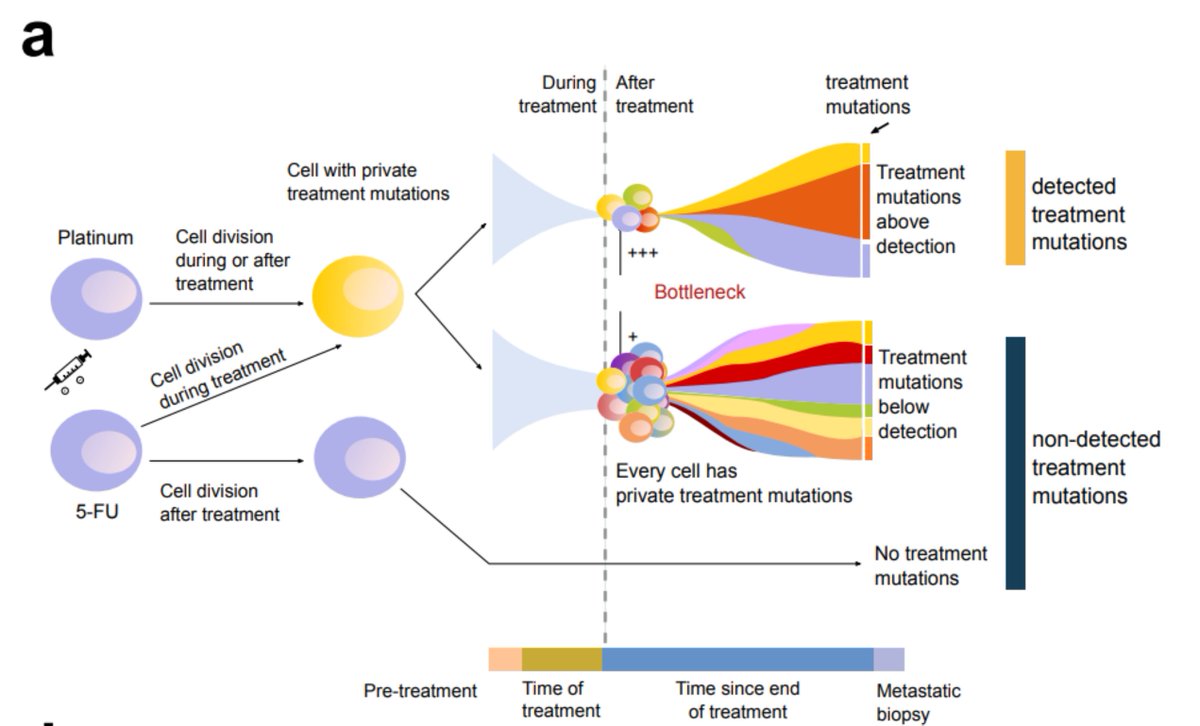
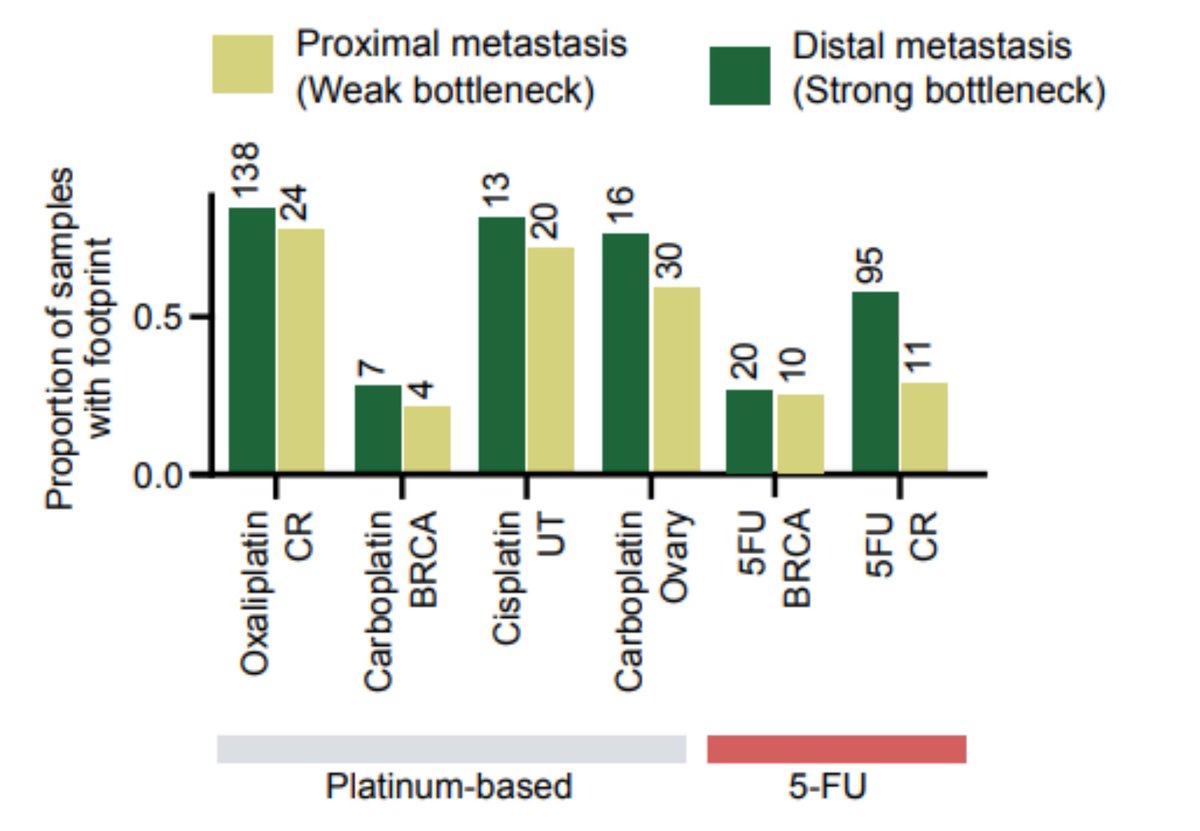
We can only observe the treatment mutations if a clonal expansion following the treatment has occurred. Inspired by recent publications ( @cncurtis), we observed that distal metastasis (with stronger bottleneck) are more likely to present a footprint, both of 5-FU and platins.
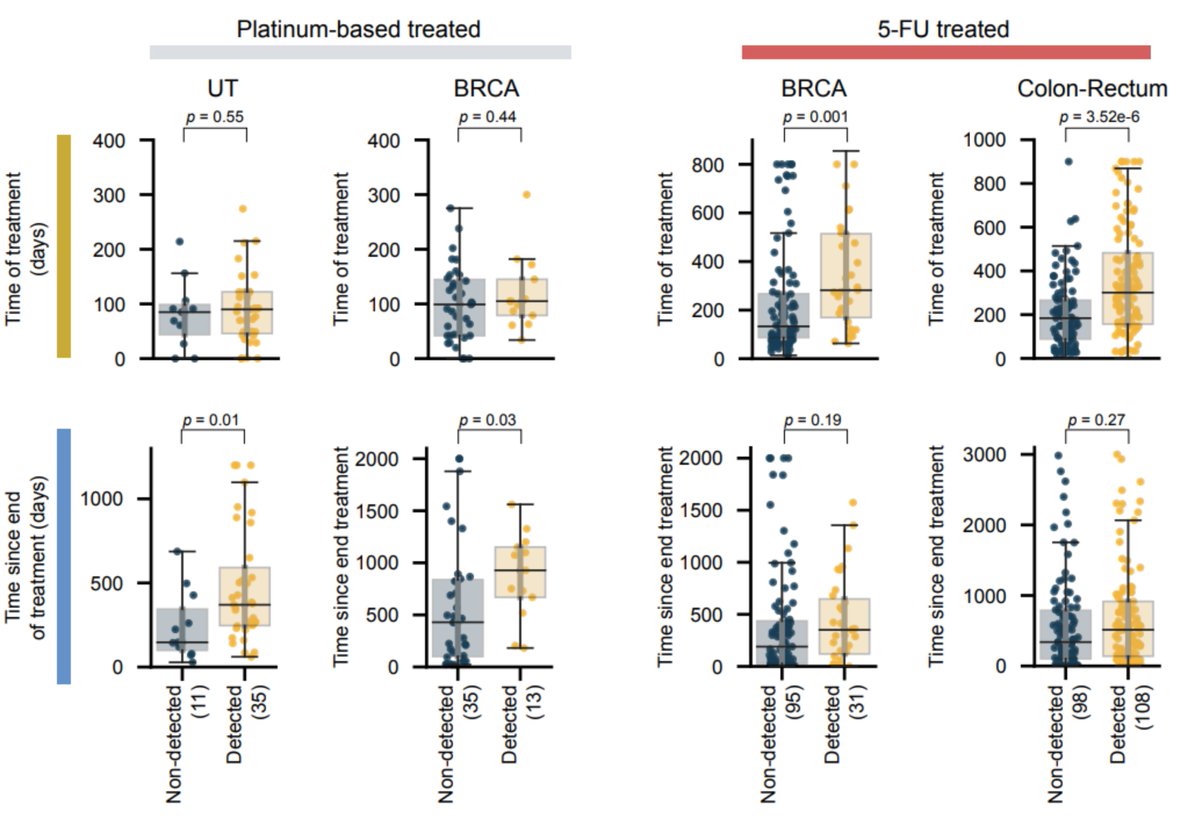
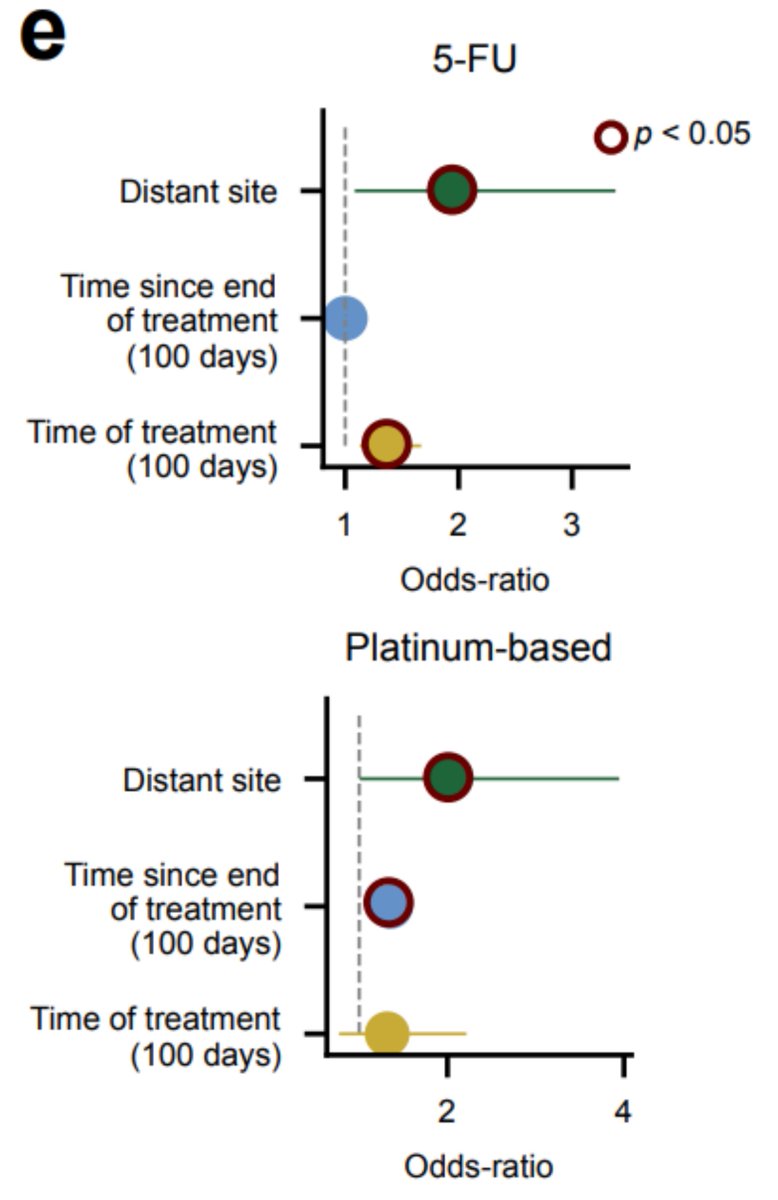
BUT There is another factor that distinguishes platinum and 5-FU exposed samples. In platinum, the time since the end of the treatment is determinant to observe the footprint, while for 5-FU, it is the duration of the treatment that seems crucial
There is another factor that distinguishes platinum and 5-FU exposed samples. In platinum, the time since the end of the treatment is determinant to observe the footprint, while for 5-FU, it is the duration of the treatment that seems crucial
 There is another factor that distinguishes platinum and 5-FU exposed samples. In platinum, the time since the end of the treatment is determinant to observe the footprint, while for 5-FU, it is the duration of the treatment that seems crucial
There is another factor that distinguishes platinum and 5-FU exposed samples. In platinum, the time since the end of the treatment is determinant to observe the footprint, while for 5-FU, it is the duration of the treatment that seems crucial
All this suggests that indeed, cells that do not replicate when exposed to 5-FU do not incorporate mutations related to this treatment, and that hematopoietic stem cells might be quiescent when they received the impact of the treatment.
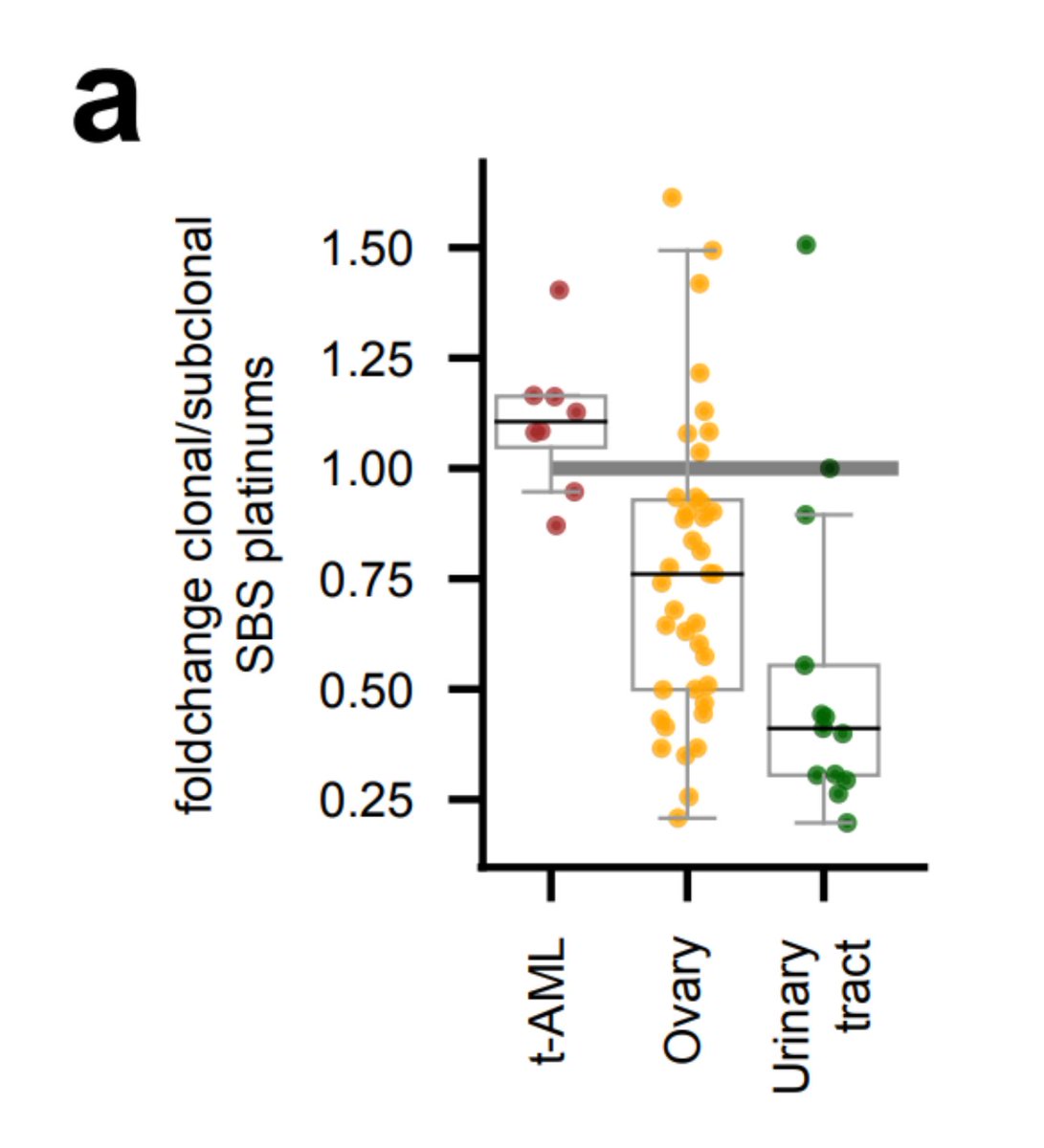
So, going back to tAMLs. Can we use the footprint to study the evolutionary constraints imposed by the chemo on hematopoietic cells? Sure!  We observed that treatment-related mutations are mostly clonal, which indicates that the treatment precedes the clonal expansion in tAMLs.
We observed that treatment-related mutations are mostly clonal, which indicates that the treatment precedes the clonal expansion in tAMLs.
 We observed that treatment-related mutations are mostly clonal, which indicates that the treatment precedes the clonal expansion in tAMLs.
We observed that treatment-related mutations are mostly clonal, which indicates that the treatment precedes the clonal expansion in tAMLs.
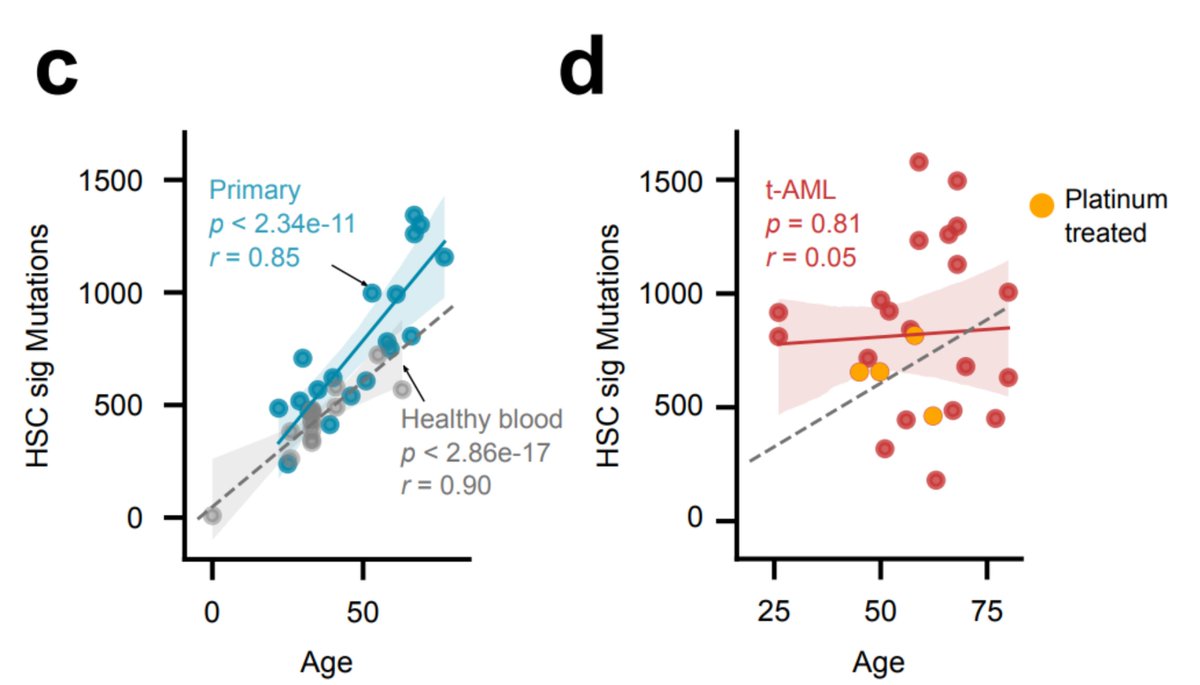
Chemos alter the developmental dynamics of the hematopoietic compartment, regardless of their mutagenic effect. In tAMLs (but not in primary AMLs) we observe a loss of the linear relationship between hematopoiesis mutations and the age of the donors seen in healthy hematopoiesis
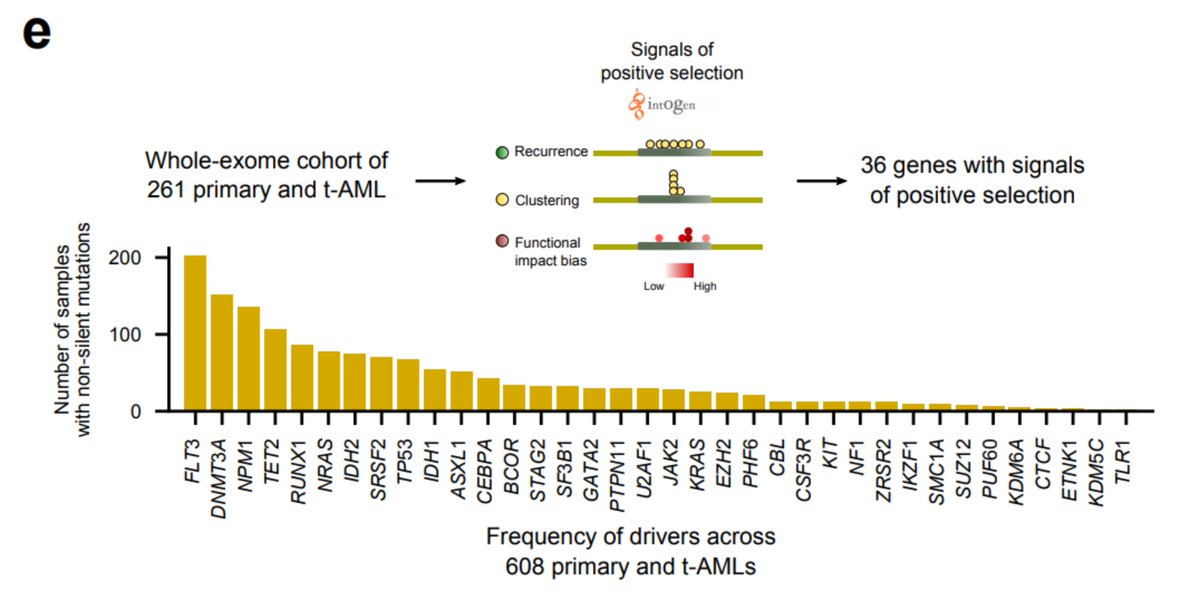
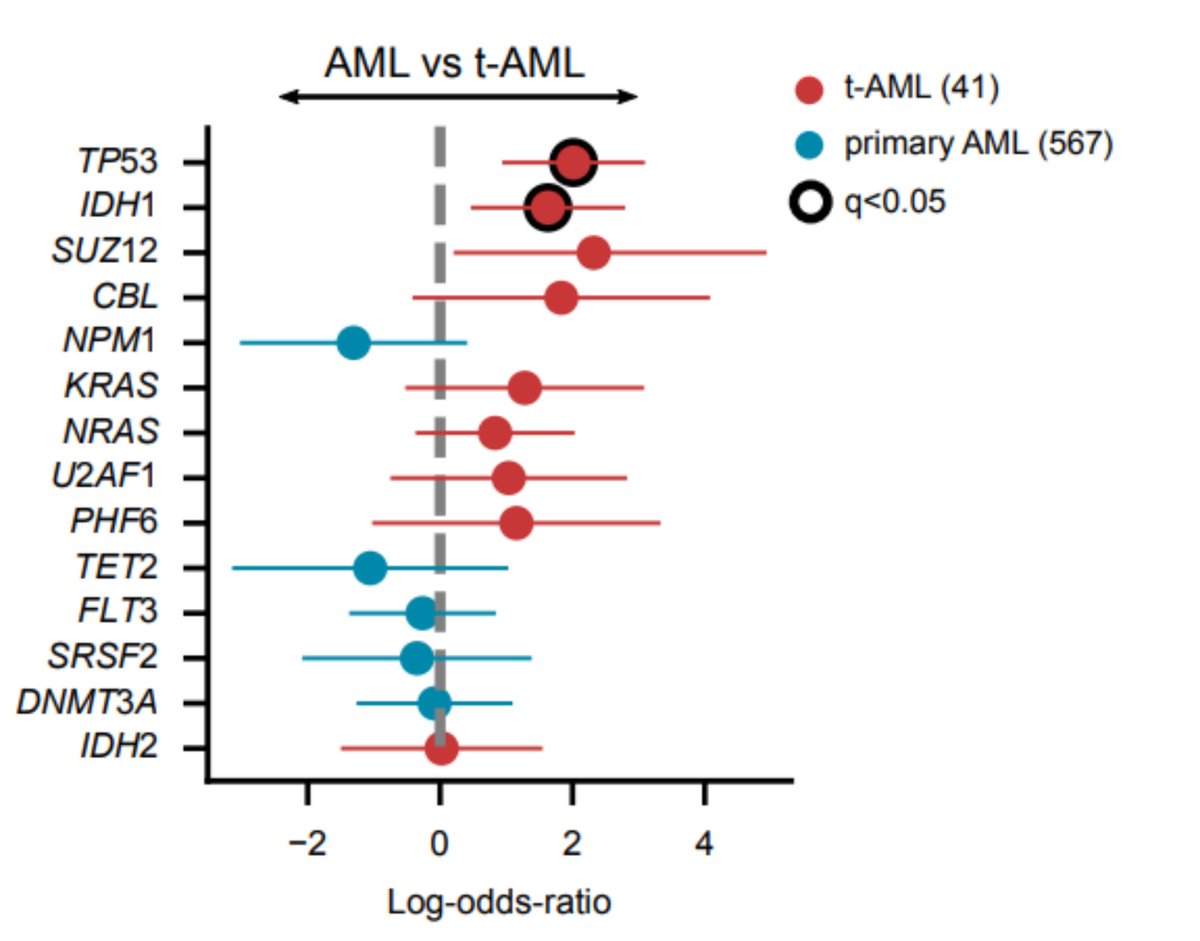
Hematopoietic cells founding the primary and tAMLs face different selective constraints, so we asked whether mutations in different genes drive both malignancies. To sort this out, we discovered genes under positive selection in #beatAML cohort
We found, as previously discovered in Wong et al. ( https://go.nature.com/2GaIZSQ ), that TP53 appears enriched in tAML cases. However, mutations of IDH1 are also significantly overrepresented in these samples.
We then focused on another treatment-related clonal expansion affecting healthy blood cells: treatment-related clonal hematopoiesis (tCH). In a recent preprint we identified the blood mutations in treated individuals and CH driver genes -read it here !
 https://twitter.com/oriol_pich/status/1320758034860253186
https://twitter.com/oriol_pich/status/1320758034860253186

 https://twitter.com/oriol_pich/status/1320758034860253186
https://twitter.com/oriol_pich/status/1320758034860253186
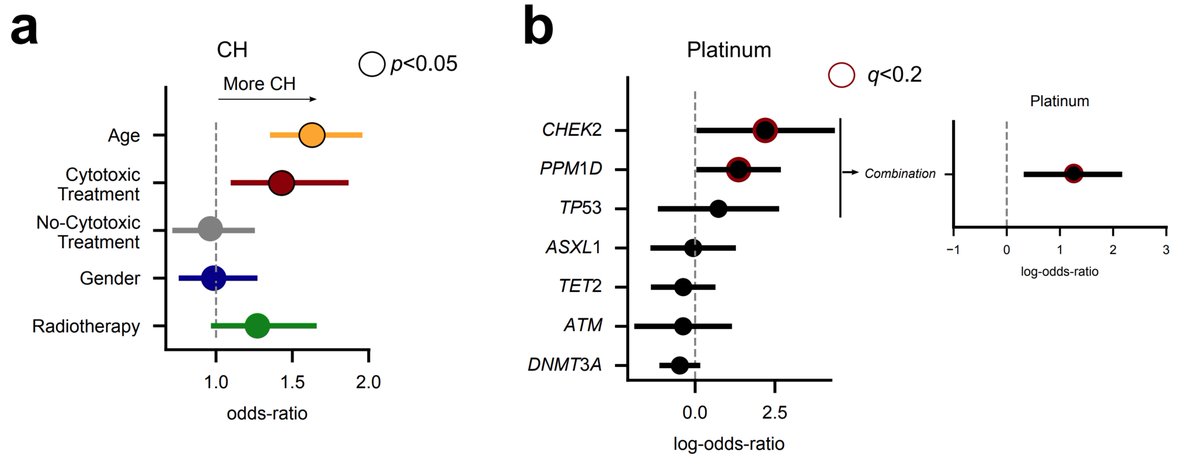
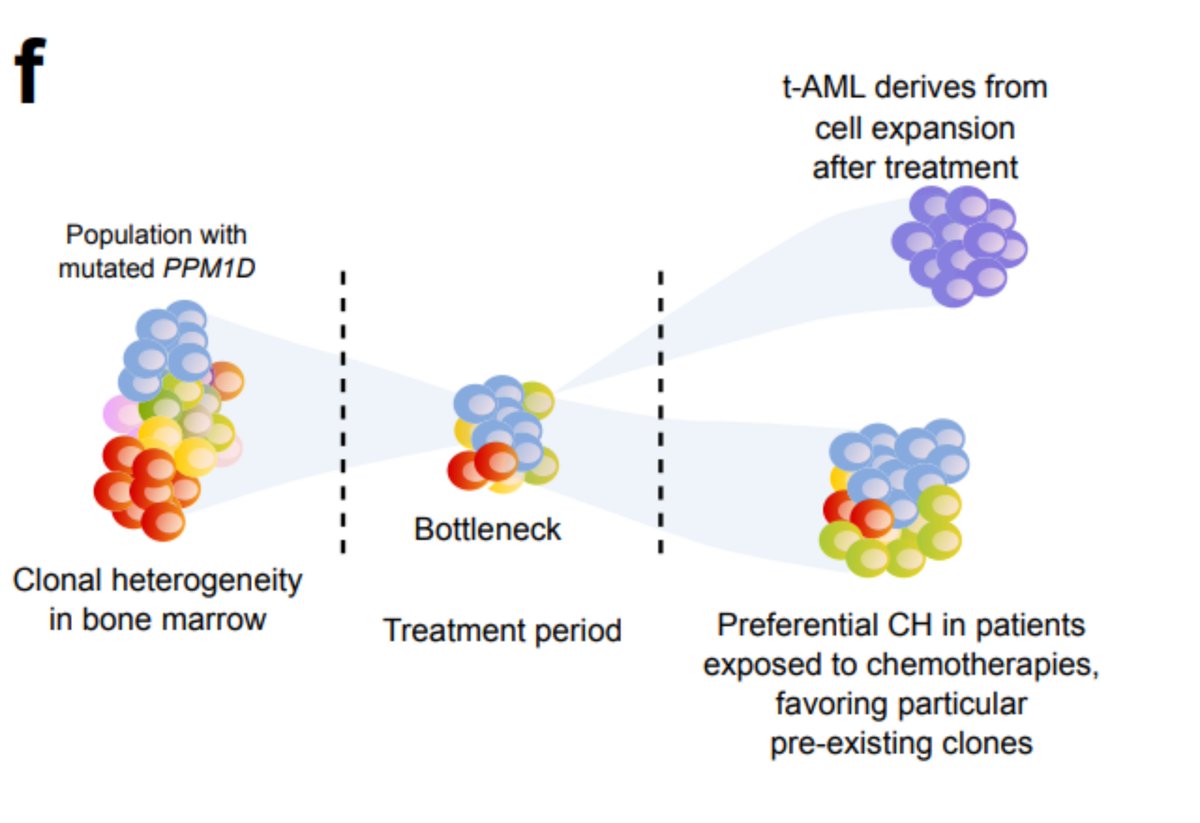
We and others ( @PapaemmanuilLab @Goodell_Lab) have shown that treatment increases the likelihood of suffering CH. Also, some CH drivers are related to a previous exposure of particular chemotherapies. This seems to be the case of platinum drugs and PPM1D, for example.
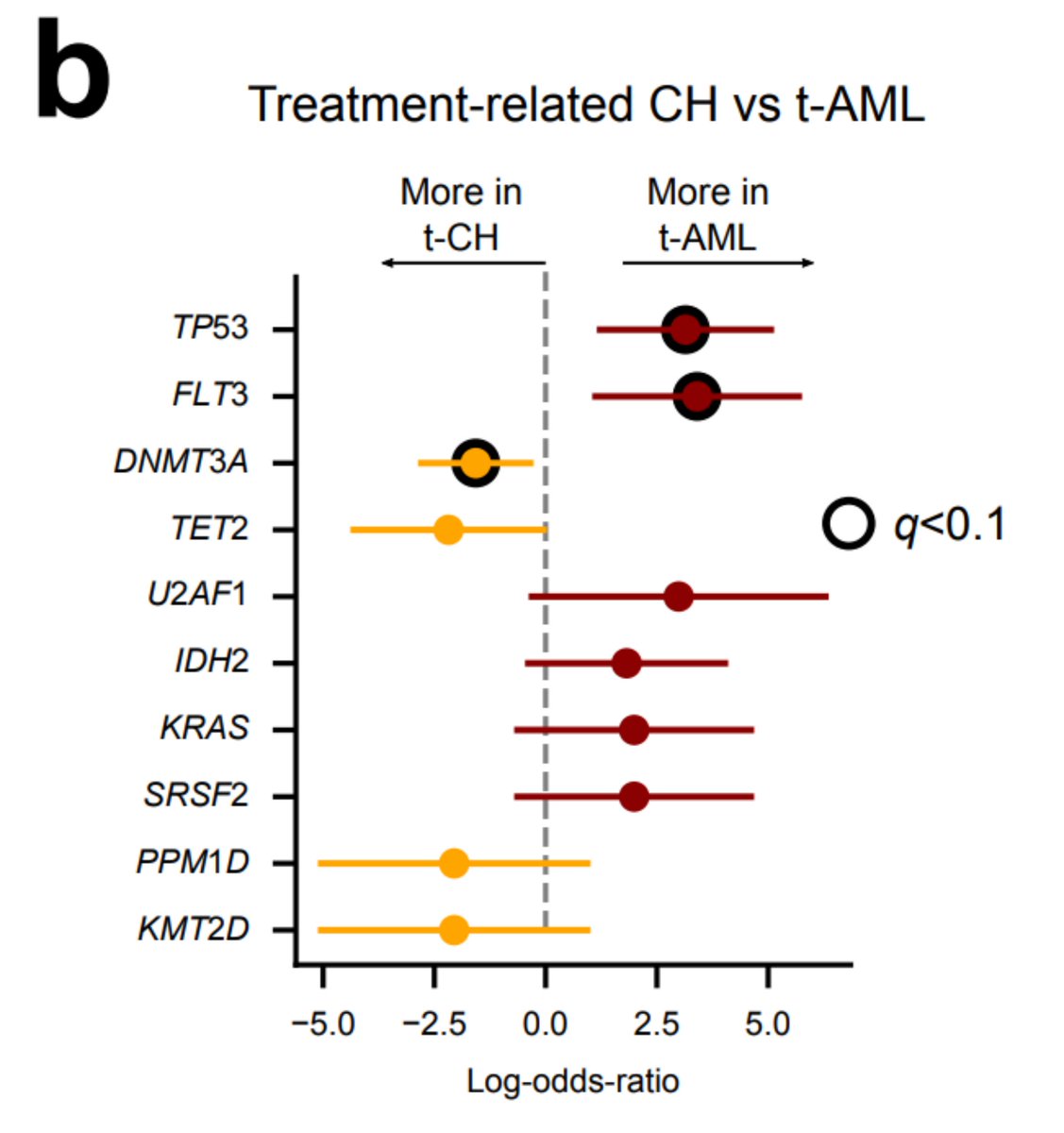
Are the most frequent driver genes of CH and tAML the same? Well, it seems there are some preferences, although it is important to note that we have fewer tAML than tCH.
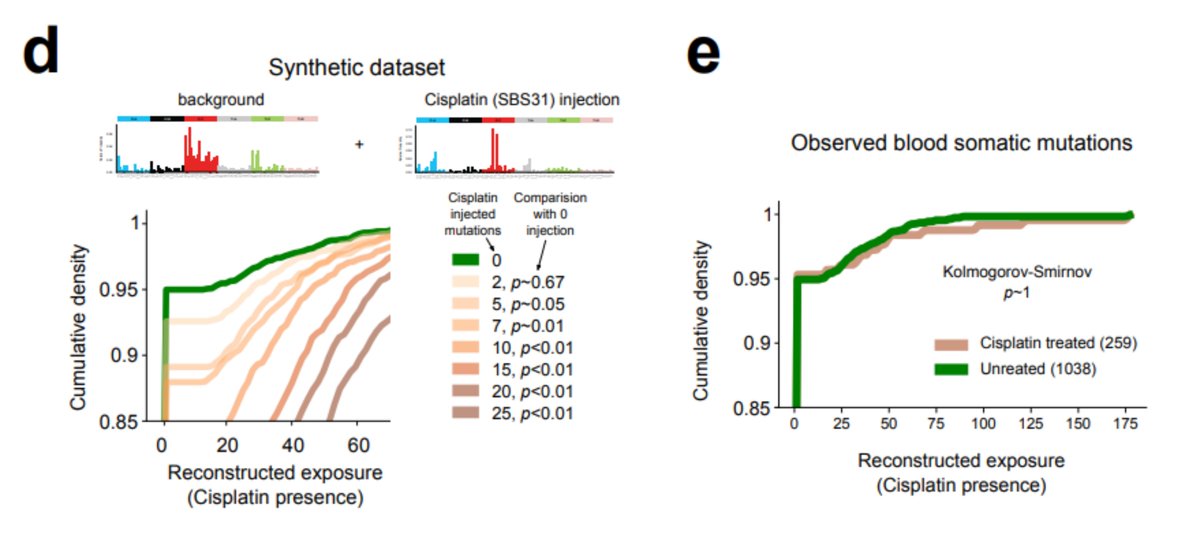
But can we identify the mutational footprints of the platinum and 5-FU among blood mutations of treated patients with CH? The standard mutational signature extraction cannot find any traces of the footprint Could this be a methodological caveat?
Could this be a methodological caveat?
 Could this be a methodological caveat?
Could this be a methodological caveat?
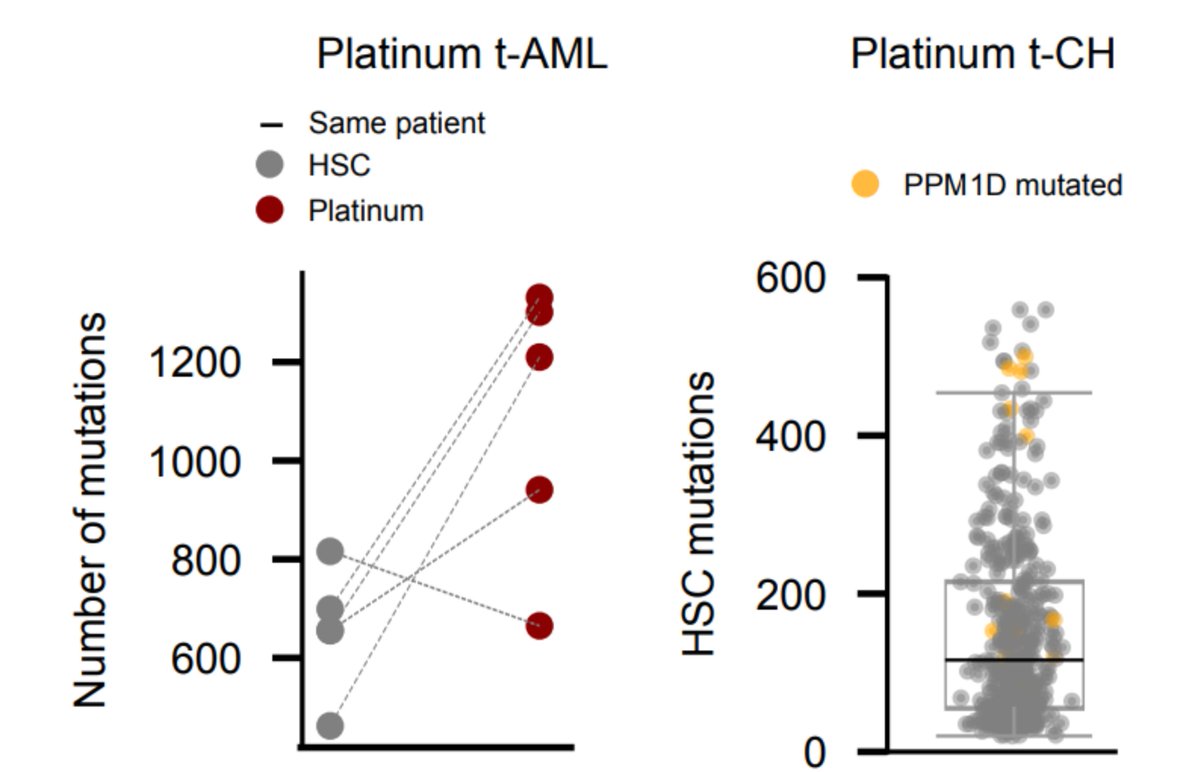
Remember we did see platinum footprint in tAMLs 
 ? We can use these samples to infer how many platinum mutations we expect given the number of hematopoiesis mutations observed in healthy blood samples.
? We can use these samples to infer how many platinum mutations we expect given the number of hematopoiesis mutations observed in healthy blood samples.

 ? We can use these samples to infer how many platinum mutations we expect given the number of hematopoiesis mutations observed in healthy blood samples.
? We can use these samples to infer how many platinum mutations we expect given the number of hematopoiesis mutations observed in healthy blood samples.
With the help of @fmuinos we devised a test, using a method by Steve Rozen, @alvinwtng, @ArnoudBoot86, that proved sensitive enough to detect the signal if at least 10 platinum-related mutations were present in the sample. Nevertheless, the signal is NOT there!
Again, keep in mind that the treatment does something - we do find PPM1D mutations preferentially in the blood of platinum-exposed donors. But this suggests a different scenario than that of tAML: the treatment favours a preexisting clone; it does not precede the clonal expansion
In summary, healthy cells exposed to some mutagenic chemotherapies receive treatment mutations; using these as barcode ( https://go.nature.com/2TyX0Nr @FrancescoMaura4), we were able to precisely time the clonal expansion underlying tAML and CH with respect to the moment of exposure.
I am really proud of this work - some of these ideas stem during one roadtrip with a great hematologist & a great friend of mine @a_cortesbu. 600 km to a three days of rock festival @VinaRockOficial and the fourth driving 600 km back to a @Metallica concert. Let there be rock !
!
 !
!

 Read on Twitter
Read on Twitter