Excited that COVID-3D has been published in Nature Genetics. This is a great piece of work from the group, but led especially @steffportelli @carlosmr12 @elston_neil and Moshe. This work was supported in particular by an @nhmrc Investigator Grant
https://www.nature.com/articles/s41588-020-0693-3
https://www.nature.com/articles/s41588-020-0693-3
There are a lot of new features for people to explore in COVID-3D (but you might need to clear your cache to make the most of them: Ctrl + Shift + R will do it  ). A bit of a thread to capture some of them:
). A bit of a thread to capture some of them:
http://biosig.unimelb.edu.au/covid3d/
 ). A bit of a thread to capture some of them:
). A bit of a thread to capture some of them:http://biosig.unimelb.edu.au/covid3d/
- Now includes mutations from >140,000 SARS-CoV-2 samples. This enabled us to explore mutational tolerance in more detail
- Some beautiful (!!!) new experimental structures. >60% more
- Information on variant and co-occurring mutations. Potential to explore compensatory effects
- Some beautiful (!!!) new experimental structures. >60% more
- Information on variant and co-occurring mutations. Potential to explore compensatory effects
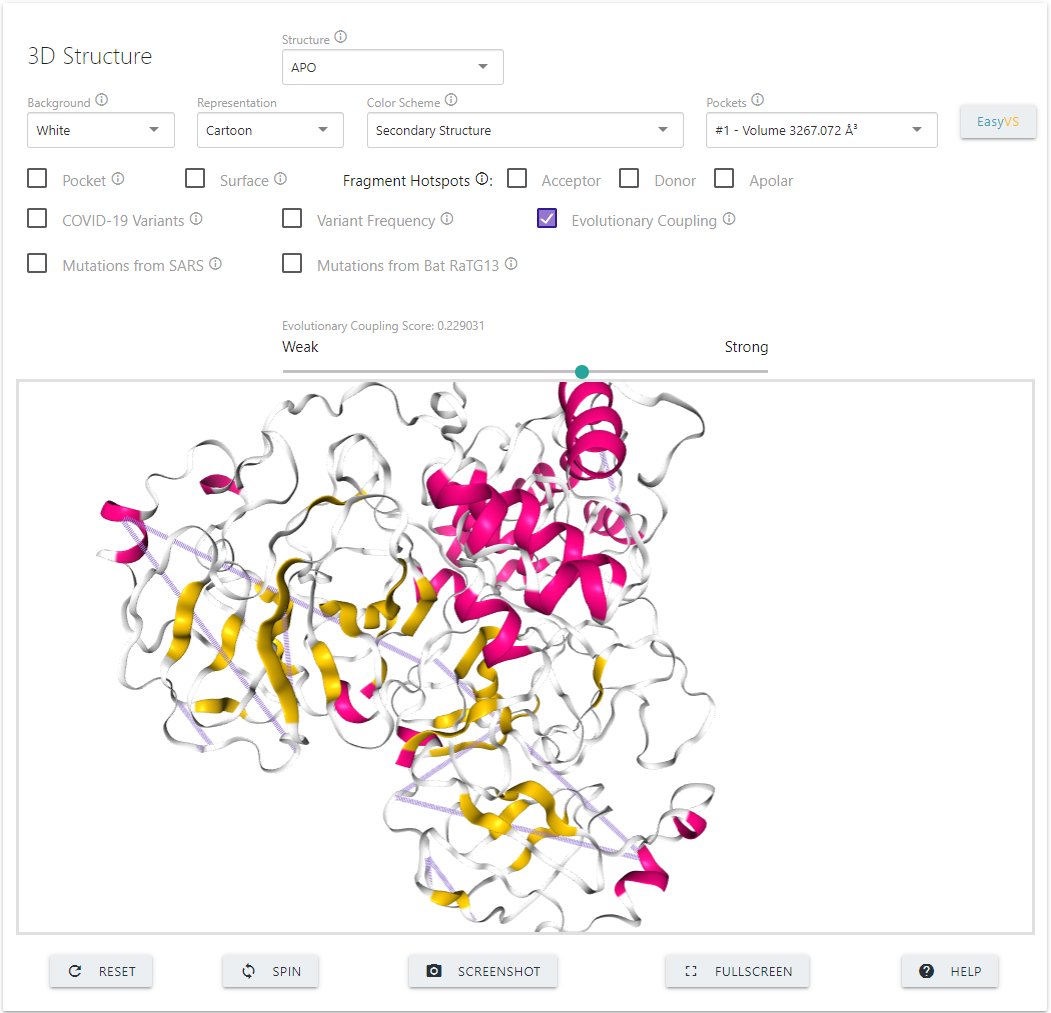
- Ability to visualise evolutionary coupling relationships to identify residues that mutate together
- Info on the mutations from SARS and Bat RaTG13 coronaviruses to enable exploration of how SARS-CoV2 jumped from Bat RaTG13, and why it is more infectious and lethal than SARS
- Info on the mutations from SARS and Bat RaTG13 coronaviruses to enable exploration of how SARS-CoV2 jumped from Bat RaTG13, and why it is more infectious and lethal than SARS
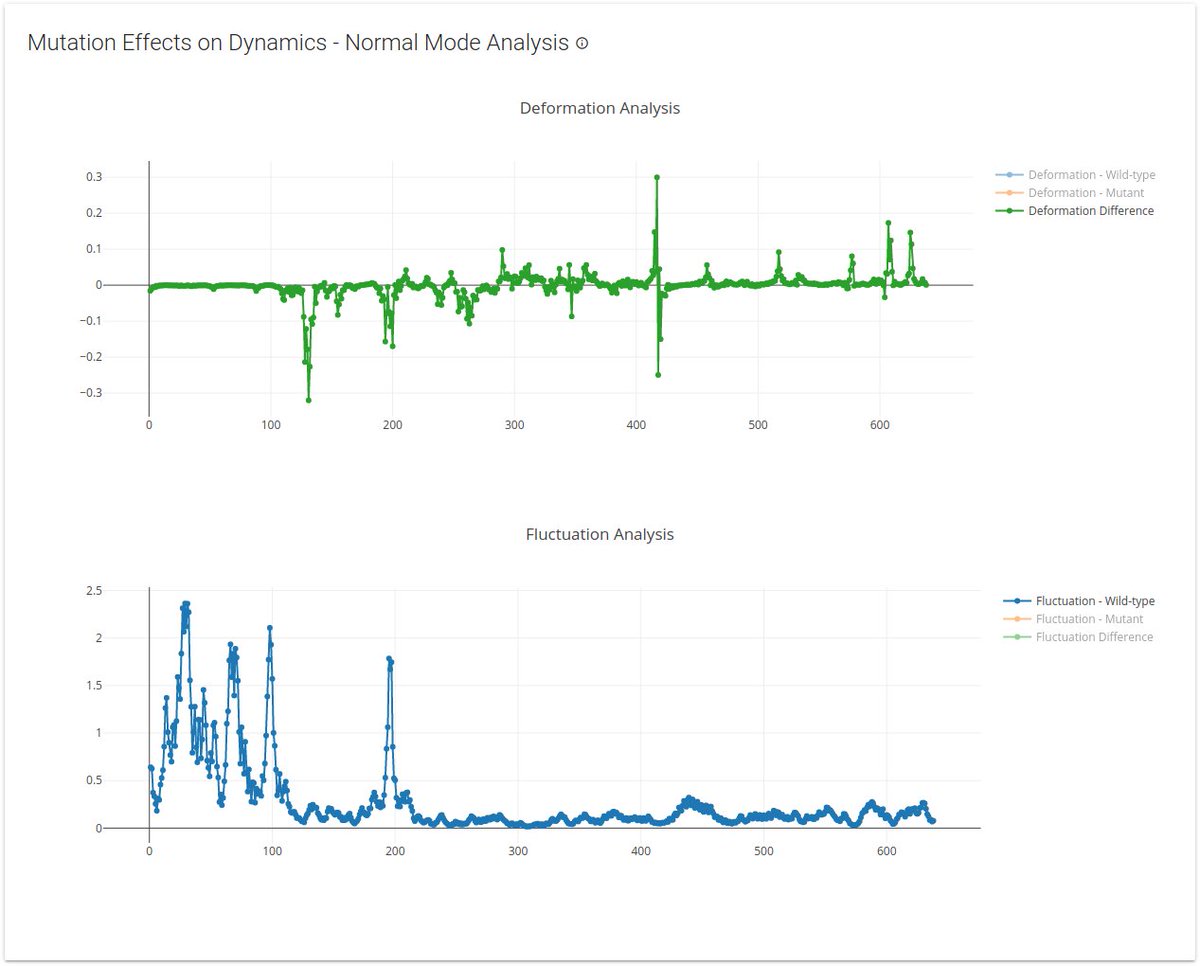
- More detailed analyses of all circulating variants are available on the mutation details page, including detailed changes in dynamics for over 20,000 variants (see how your mutation is likely to affect protein structure & function via changes in flexibility & conformation )
- The ability to launch virtual screening jobs to any pocket of interest (to enable easy and rapid identification and hypothesis of likely inhibitors that bind to a pocket under greater selective pressure- including screening for already approved drugs)
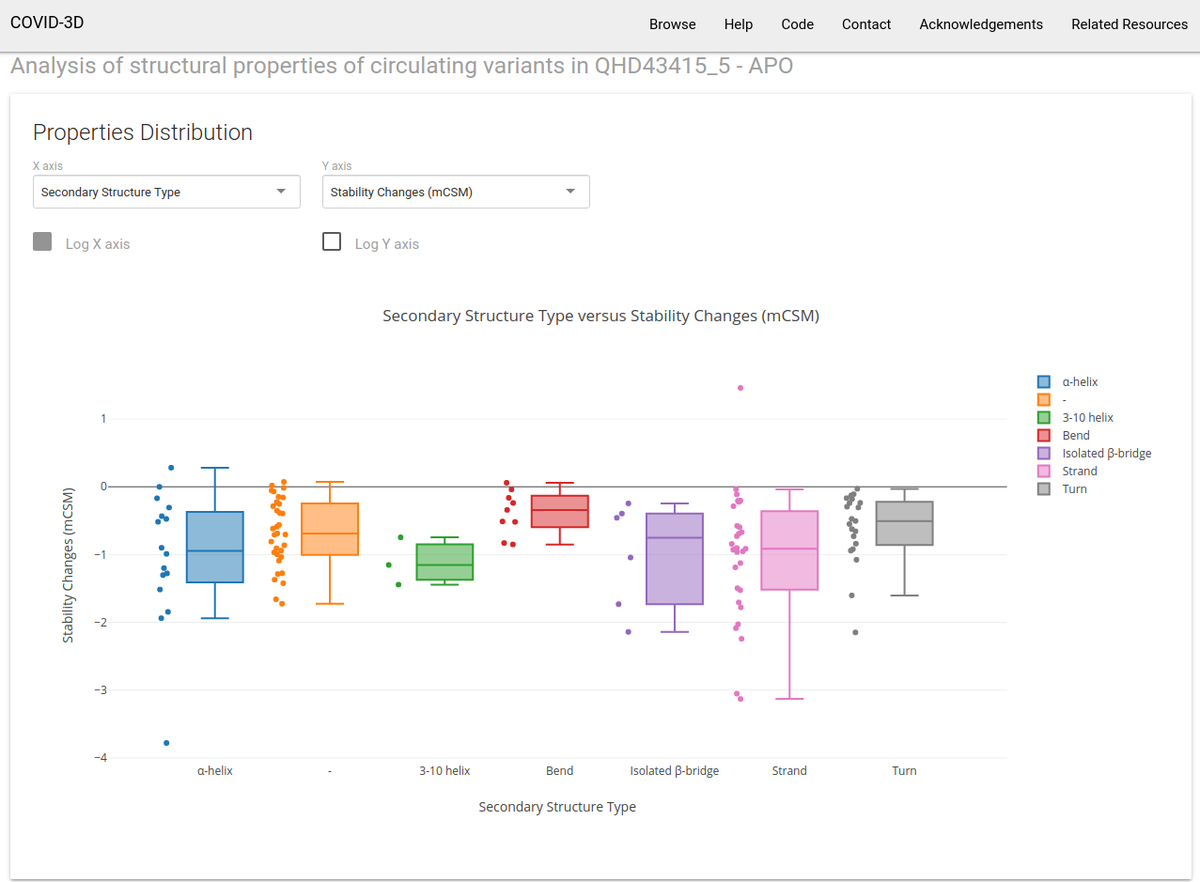
- An online tool to explore distributions and correlations of calculated properties of circulating variants (shown in Figure S7; enabling users to generate hypotheses around the relationships between biological and potential molecular and structural mechanisms)
- In silico saturation mutagenesis so that users can explore the structural consequences of genetic variants we haven't seen in circulating strains yet
- The ability to upload lists of your own variants to explore structurally and functionally here: http://biosig.unimelb.edu.au/covid3d/user_input
- The ability to upload lists of your own variants to explore structurally and functionally here: http://biosig.unimelb.edu.au/covid3d/user_input
A special thanks to @thirdreviewer , who while wasn't our strongest supporter, pushed us to make the resource even more powerful.
For those interested in the evolution of papers, the link to the original BioArxiv is: https://www.biorxiv.org/content/10.1101/2020.05.29.124610v1

 Read on Twitter
Read on Twitter