Thrilled to see our collab (driven by the fantastic Inga) in @eLife at last! Whether you're a noob, novice or grandmeister in biophysics and thermodynamics, this paper will appeal to you:
https://elifesciences.org/articles/57264
Read on for a short thread of highlights... (1/15)
https://elifesciences.org/articles/57264
Read on for a short thread of highlights... (1/15)
Molecular interactions underpin Biology. Understanding the thermodynamics (rates of association/dissociation, Kds) is crucial to modeling, understanding and manipulating these interactions. Though powerful, it's easy to get these wrong
(2/15)
(2/15)
How wrong? We scored equilibrium Kd measurements of RBP-RNA interactions from 100 papers on 2 quality criteria:
1. was incubation time varied (i.e., was thermodynamic equilibrium shown)?
2. was concentration varied (i.e., could Kd have been affected by titration)?
(3/15)
1. was incubation time varied (i.e., was thermodynamic equilibrium shown)?
2. was concentration varied (i.e., could Kd have been affected by titration)?
(3/15)
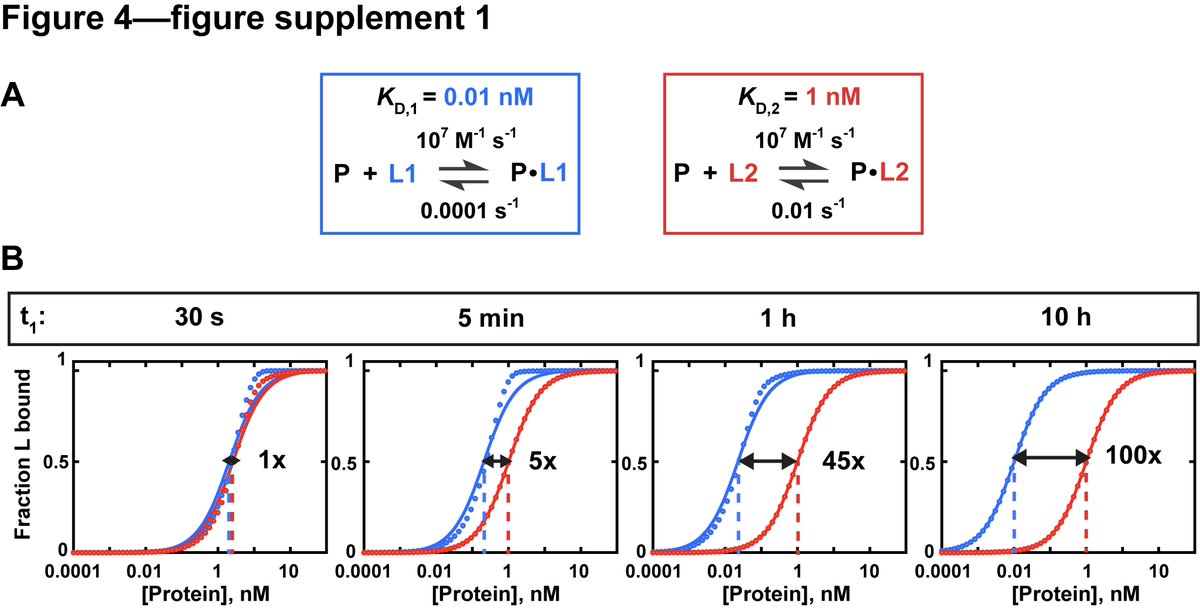
It can take anywhere from millisec to days for equilibrium to be reached (depending on the binding affinity, Kd) Why is this important? Not waiting for equilibrium to be reached can result in incorrect estimates of the Kd.
(5/15)
(5/15)
Most papers reported an incubation time of 1hr or less. But if you don't wait long enough, affinities that are off by a 100-fold can appear to be the same. E.g., CRISPR Cas12a complex has such a low koff (dissociation rate) that it needs to equilibrate for >100 hrs!
(6/15)
(6/15)
Our model RBP, Puf4, equilibrates within 30 min at 25C but at 0C needs at least 4.5 hr to equilibrate fully. We would not have been able to figure this out unless we varied incubation times. (7/15)
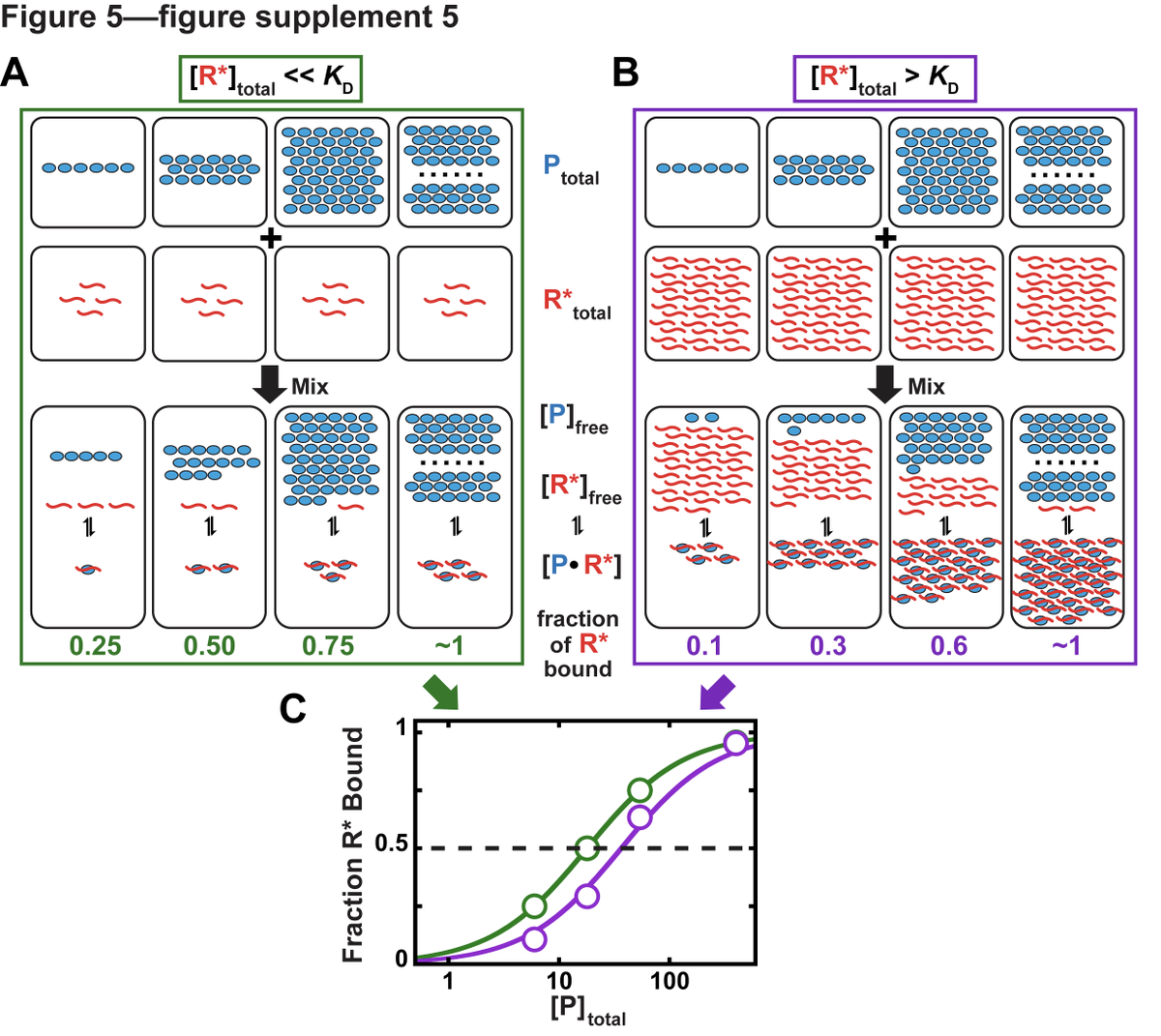
Second, it's important to ensure that you're not in a titration regime where ligand [R] >> Kd. At high [R], [P] gets depleted from solution such that [P]free ~ [P]total is no longer true. Kds cannot be measured when [R] is in vast excess over Kd (tho workarounds exist) (8/15)
Although 73% of papers use an appropriately low concentration of the ligand, only 5% of studies actually varied ligand concentration. 30% of reported Kds could have been affected by titration. (9/15)
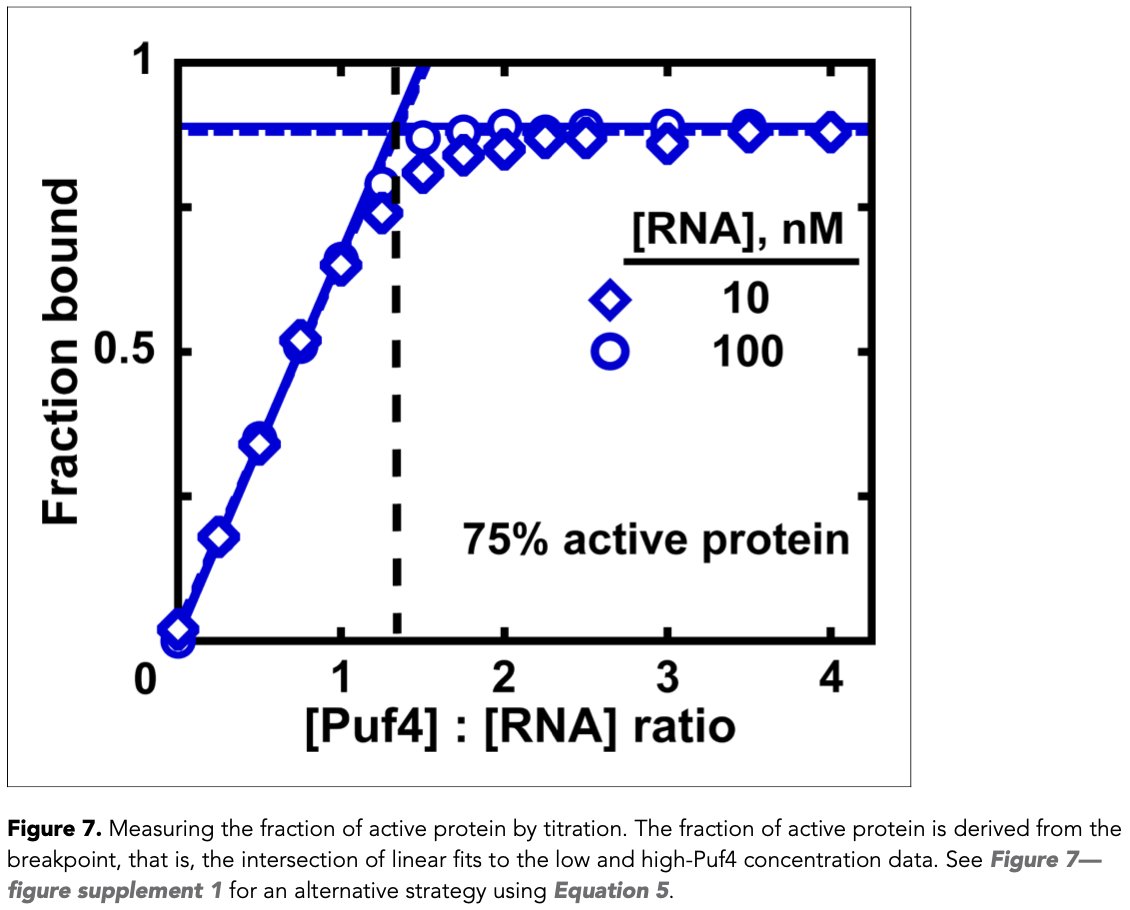
One place where titration regime is actually good is when you are measuring fraction of active protein in your protein prep. The fraction of active protein is derived from the intersection of linear fits to the low and high-protein concentration data (10/15)
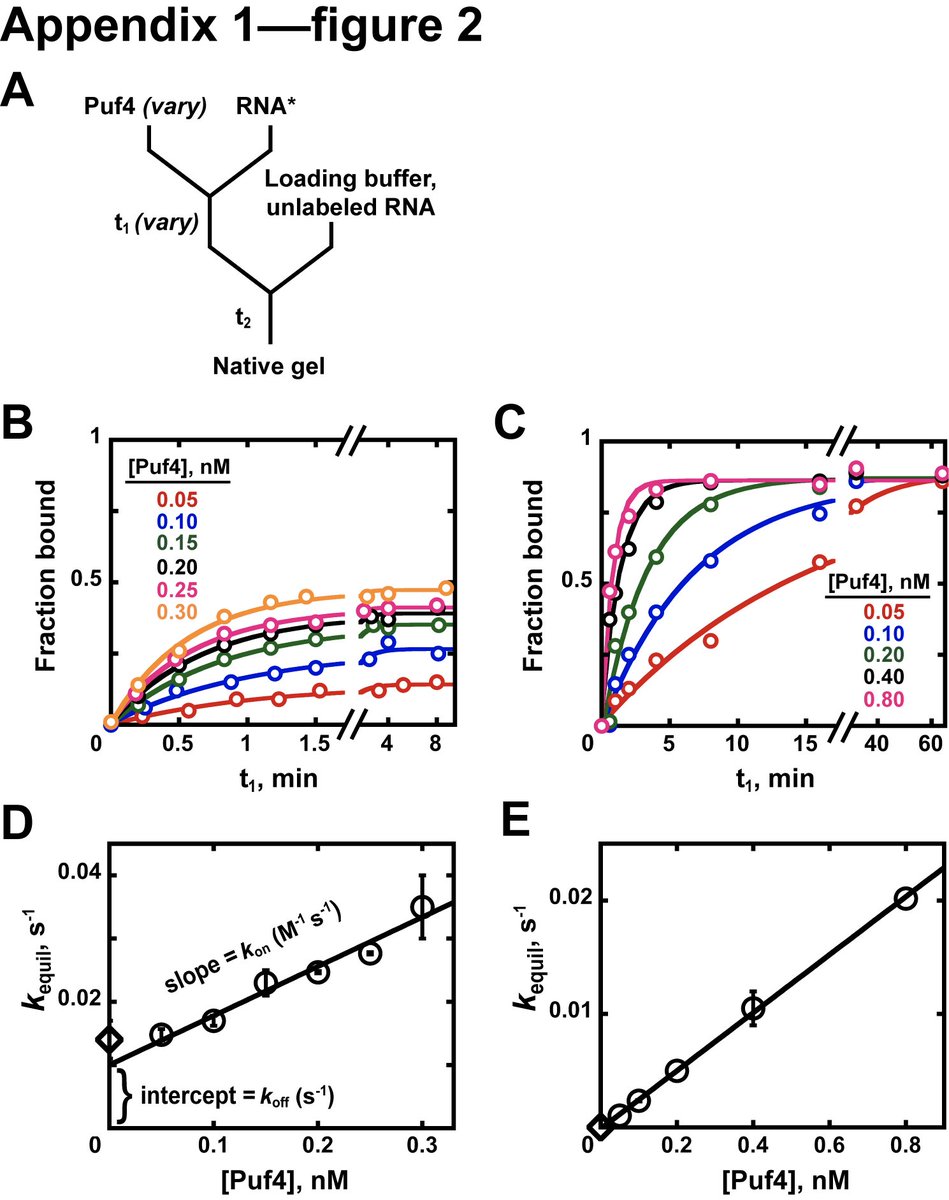
Where possible, Kd measurements should also be validated by an orthogonal method. Since Kd = koff/kon, kinetic measurements of association (kon) and dissociation (koff) rate constants provide an easy way to confirm Kd
(Figure panels show koff, kon at 25C and 0C) (11/15)
(Figure panels show koff, kon at 25C and 0C) (11/15)
Immunoprecipitation and pulldown assays are often interpreted in terms of 'binding' or 'no binding' - but observing binding in these assays is a complex function of the experimental components and conditions and is a lot more nuanced! (12/15)
2 RBPs with same Kds to RNA can be categorized completely differently because of the differential effects from the RNA pulldown procedure on the kinetic parameters (kon, koff) of the proteins binding to RNA. [See paper for more details] (13/15)
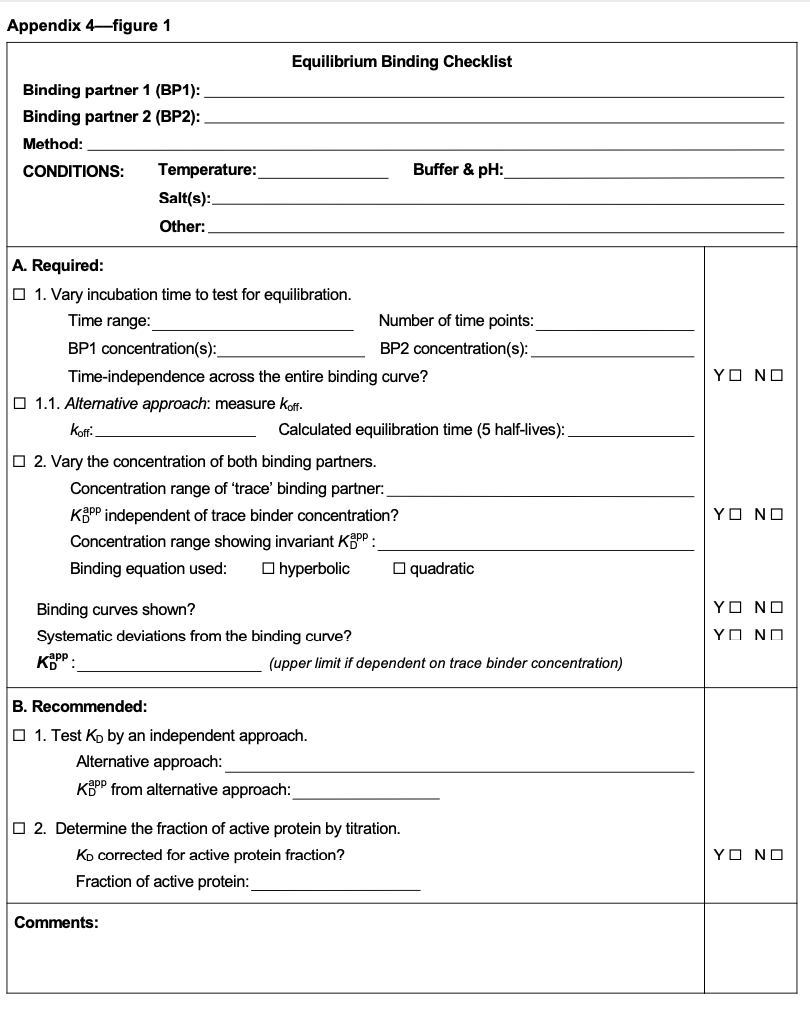
We hope that these illustrations and guides help clarify the concepts around measuring thermodynamics parameters. To aid in the actual experiments, we've also developed a convenient Equilibrium Binding Checklist, that we hope will be widely adopted by the community
(14/15)
(14/15)
Please make sure to read through the notes and appendices for lots of other interesting and useful nuggets. And don't hesitate to reach out with feedback/questions. Happy thermodynamicsing! (15/15)
P.S. In Tweet #1 of the thread, I meant to say "Grandmaester". TIL that the other thing is a Guitar Amplifier apparently. Le sigh.

 Read on Twitter
Read on Twitter




![Second, it's important to ensure that you're not in a titration regime where ligand [R] >> Kd. At high [R], [P] gets depleted from solution such that [P]free ~ [P]total is no longer true. Kds cannot be measured when [R] is in vast excess over Kd (tho workarounds exist) (8/15) Second, it's important to ensure that you're not in a titration regime where ligand [R] >> Kd. At high [R], [P] gets depleted from solution such that [P]free ~ [P]total is no longer true. Kds cannot be measured when [R] is in vast excess over Kd (tho workarounds exist) (8/15)](https://pbs.twimg.com/media/EfzLHTvUMAA24m0.jpg)