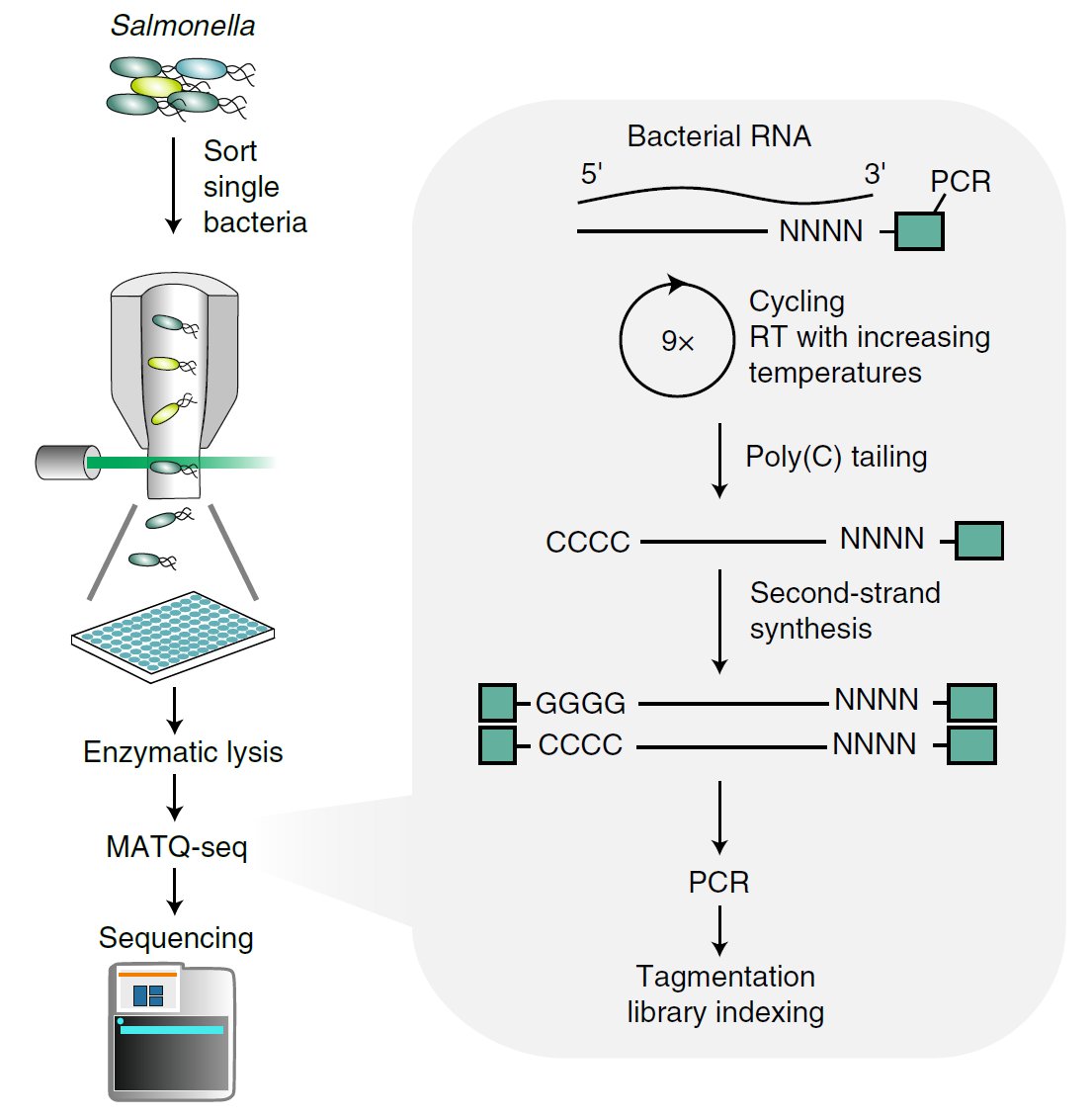
It is a great honor to present our last study published today in @NatureMicrobiol. We pushed the limit of single-cell RNA-seq to report the physiology of a single bacteria, using Salmonella and Pseudomonas as pathogens (1/5). Link: https://www.nature.com/articles/s41564-020-0774-1
A real challenge to capture bacterial RNAs! They do not have a poly(A) tail and are enclosed behind a thick cell wall in femtogram levels. We adopted a recently developed single-cell RNA-seq protocol called MATQ-seq to capture all RNA classes… (2/5)
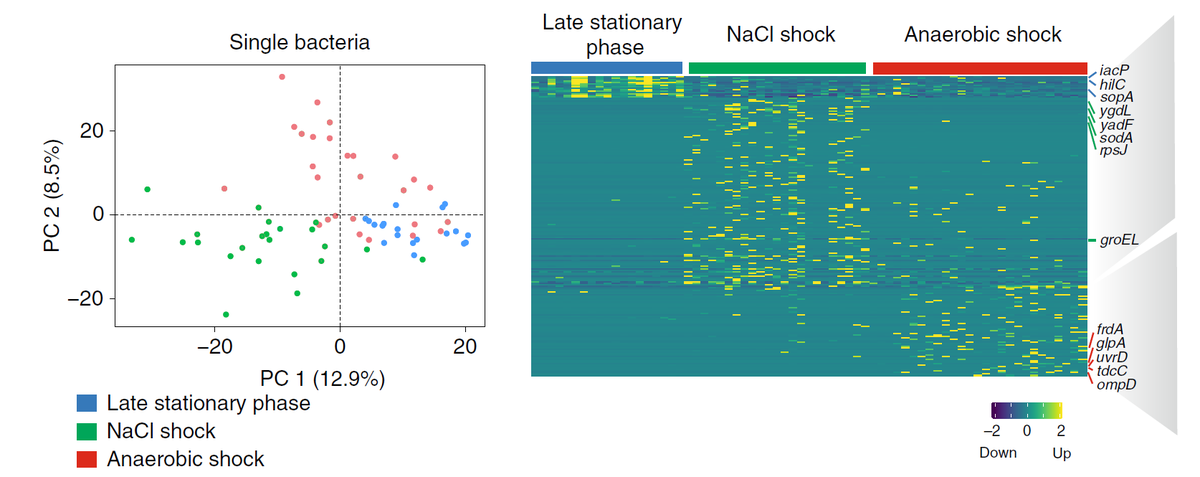
… and we could successfully delineate Salmonella physiology grown under different stress conditions (3/5).
We believe that it will be incredibly important to understand the physiology of individual bacteria if we want to tackle persisters and the microbiome. I am very proud of my students @FabianImdahl and Ehsan Vafadarnejad. A collaboration with Jörg Vogel @Hiri @Helmholtz_HZI (4/5).
To read the paper, it is easy: https://rdcu.be/b6jH1
(5/5)
(5/5)

 Read on Twitter
Read on Twitter