So happy that our paper determining the structure of Parkinson’s Disease linked LRRK2 in situ is now in print. Since I didn’t described it when we posted it to bioRxiv, here it goes.. 1/n http://disq.us/t/3qz3uag
~10% of Parkinson’s disease cases are linked to a genetic cause, with mutations in the LRRK2 gene being the most common cause of PD in this group. LRRK2 was discovered 16 years ago, and while an obvious drug target, determining its structure has been incredibly hard. 2/n
Efforts included sending the protein to the IIS to crystallize under zero gravity (no dice). It’s a large protein, notably difficult to work with – some call it a sticky business. 3/n https://www.nasa.gov/mission_pages/station/research/news/parkinsons-research
Susan Taylor, kinase extraordinaire, introduced me to LRRK2. What fascinated me about this protein is it has a GTPase AND a kinase – the 2 greatest signaling domains in cell biology together in one protein! PD mutations are localized 2 these domains, and the rest is made of.. 4/n
.. protein-protein interaction domains (hence the stickiness). Several mutants of LRRK2 were observed to bind and decorate microtubules when overexpressed, so we used this as an assay to determine the structure #insitu. 5/n
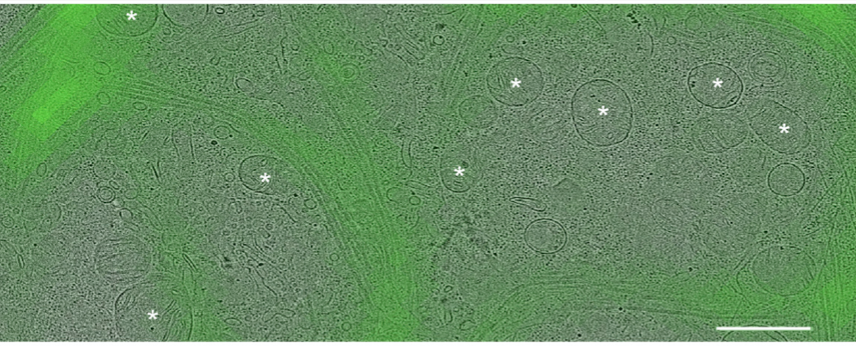
First, we used #CLEM to locate YFP-tagged #LRRK2 in HEK cells with the I2020T PD mutation. We selected for MTs decorated with #LRRK2. Then, we used #cryoFIB milling and #cryoET to image the MTs. 6/n
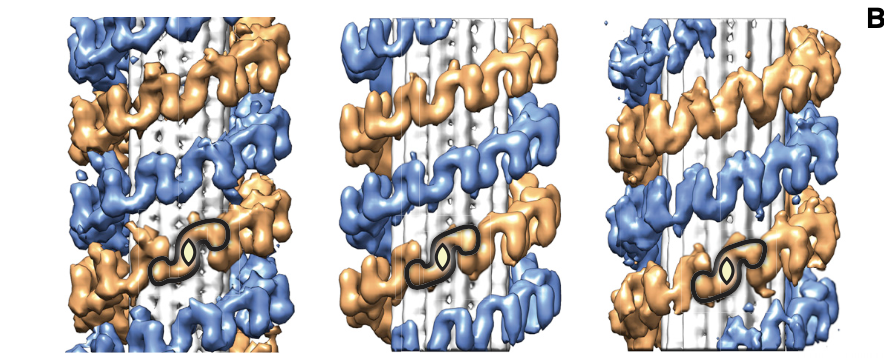
We found that even though most cells in culture have predominantly 13-protofilament MTs, LRRK2-decorated MTs had preferentially 11 and 12 protofilaments, and that LRRK2 formed a right-handed double helix around the left-handed microtubules! 7/n
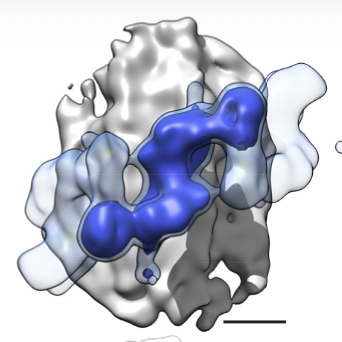
Next, we found that the repeating unit in the LRRK2 helix was a LRRK2 dimer, and while the helix was regular, cells are messy test tubes and symmetry imperfect, so we isolated subtomograms of dimers to increase the resolution. This resulted in a 14-A map of MT-bound LRRK2. 8/n
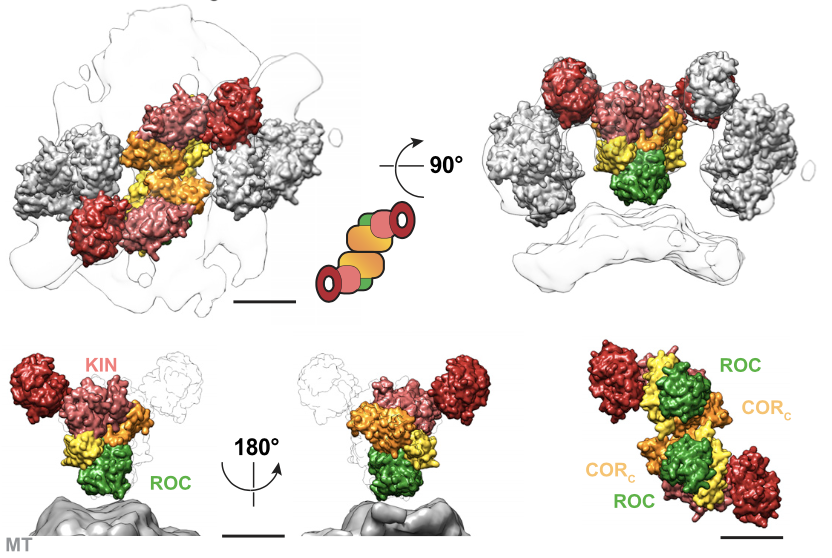
From here, did integrative modeling using IMP. From 3M initial models, we unveiled the molecular architecture of MT-bound LRRK2, that shows the kinase facing the cytoplasm and in close proximity to the GTPase, that faces the MT. 9/n
Homotypic interactions between the WD40 and COR domains form the filaments. PD LRRK2 mutants show kinase hyperactivity. Our data suggests an an active-like kinase conformational state of LRRK2 bound to MTs. 10/n
To our knowledge, it's the highest resolution structure of a eukaryotic protein in situ not previously determined by in vitro methods. If you wonder, we think that w/ #cryoET and #StA it can go to higher res, but had exciting collaborative results in the pipeline (Stay tuned) 11/n
Most important, this work was done by wonderful people who make me feel honored to have a lab! @R_Buschauer started the project as an MS student in the lab, continued by @ReikaWatanabe2 who became a jack of all trades and saw this project through.. 12/n
@janboeh, also as a MS had to invent a lot of creative ways to analyze the data to figure out #StA of the helices and dimers. @MartinaAudagno3 and @LaskerKeren did the integrative modeling. Tsan-Wen Lu and Daniela Boassa contributed cell biology of the G2385R LRRK2 mutant. 13/n
We are very thankful for support from several agencies, with a special shoutout to @MichaelJFoxOrg that seeded this project and has built a great collaborative network. 14/n
Apologies if you can't access our work behind the paywall. Please see the bioRxiv version or download the accepted version from our website. I will post the author link when provided by @CellCellPress when the paper appears in the print issue 15/n https://www.biorxiv.org/content/10.1101/837203v1
We are now in love with LRRK2  . A lot of exciting new directions and opportunities in our lab and our collaborators' labs that I'll discuss in upcoming threads. Thanks for reading! FIN
. A lot of exciting new directions and opportunities in our lab and our collaborators' labs that I'll discuss in upcoming threads. Thanks for reading! FIN
 . A lot of exciting new directions and opportunities in our lab and our collaborators' labs that I'll discuss in upcoming threads. Thanks for reading! FIN
. A lot of exciting new directions and opportunities in our lab and our collaborators' labs that I'll discuss in upcoming threads. Thanks for reading! FIN

 Read on Twitter
Read on Twitter