Selfing is the safest sex in C. tropicalis https://bit.ly/wgate2 the unexpectedly controversial fruit of years with @wormsrock Hannah Seidel @ecandersen John Yuen @c_braendle @Lewis17Stevens @jacjacs_ et al. It's common knowledge some worms, given the choice, prefer to fuck ...
themselves. None more so than C. tropicalis (FKA sp. 11), the most recently discovered* of 3 Caeno sp. in which females figured out how to make sperm. (*le chevalier d'histoire naturelle des nématodes, Marie-Anne Felix). We set out to study recombination and population genetics.
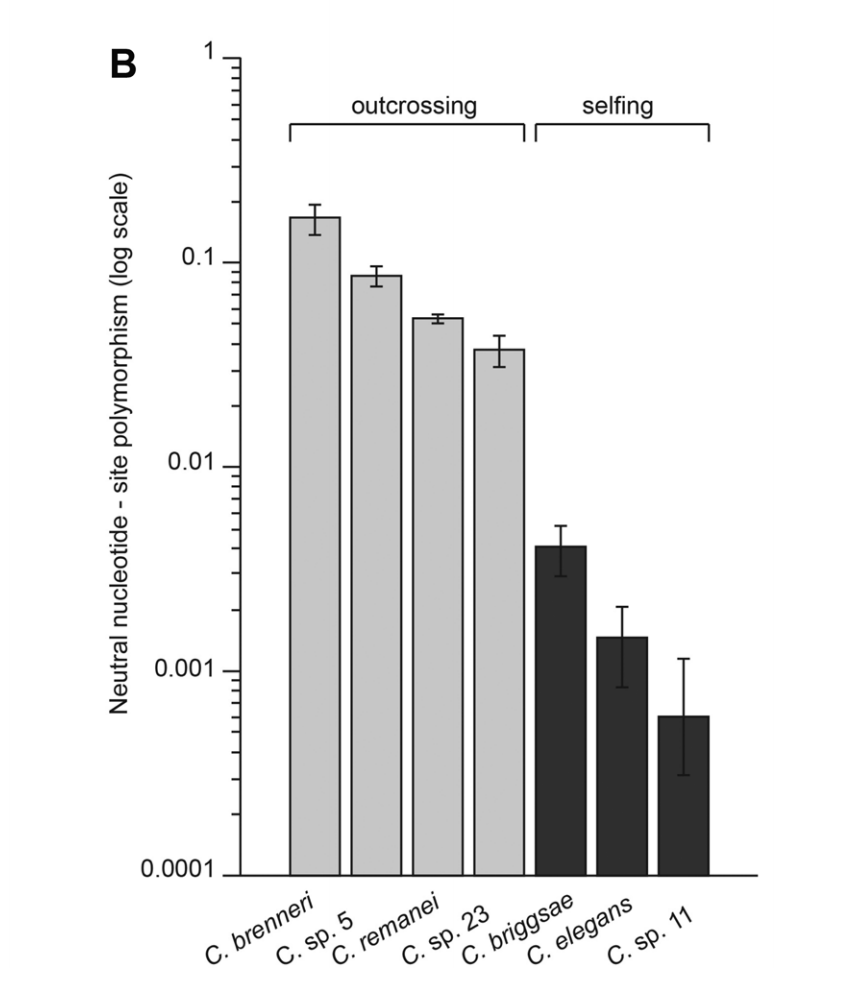
From @c_braendle @asherdcutter et al. we suspected tropicalis had little genetic diversity, and knew it was especially prone to locale-structured outbreeding depression. From @Fierst_Lab @granex et al. we knew genome shrinkage was most extreme of the 3 selfers.
We made RILs and a chromosome-scale reference genome from PacBio + linkage data, and surveyed global diversity. We found strong population structure, and really strong heterogeneity in genetic diversity (from short-read data. More PacBio showed this to be a huge underestimate).
Theory from @magnusnordborg and Charlesworths provides one potential explanation: strong balancing selection, which may be particularly easy to detect and localise in partially selfing yet large Ne species. Many divergent loci are on the scale of a few genes (in the reference).
Recent work on elegans (and briggsae) showing enrichment of ecologically-relevant genes, QTL, and gene expression in divergent regions makes this very plausible https://bit.ly/wgate1 . Other things seem also to be at play, in tropicalis especially https://bit.ly/burgaetal
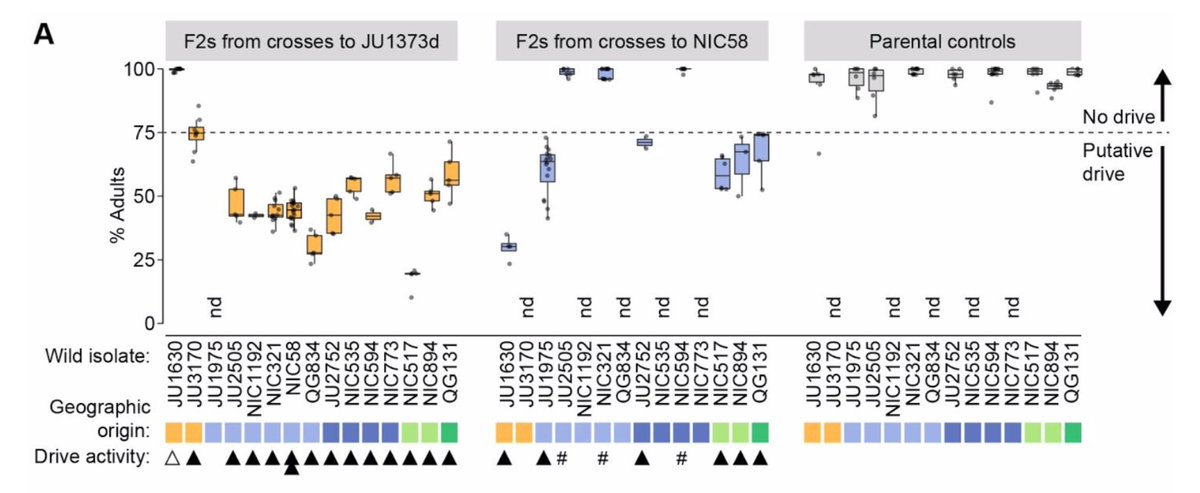
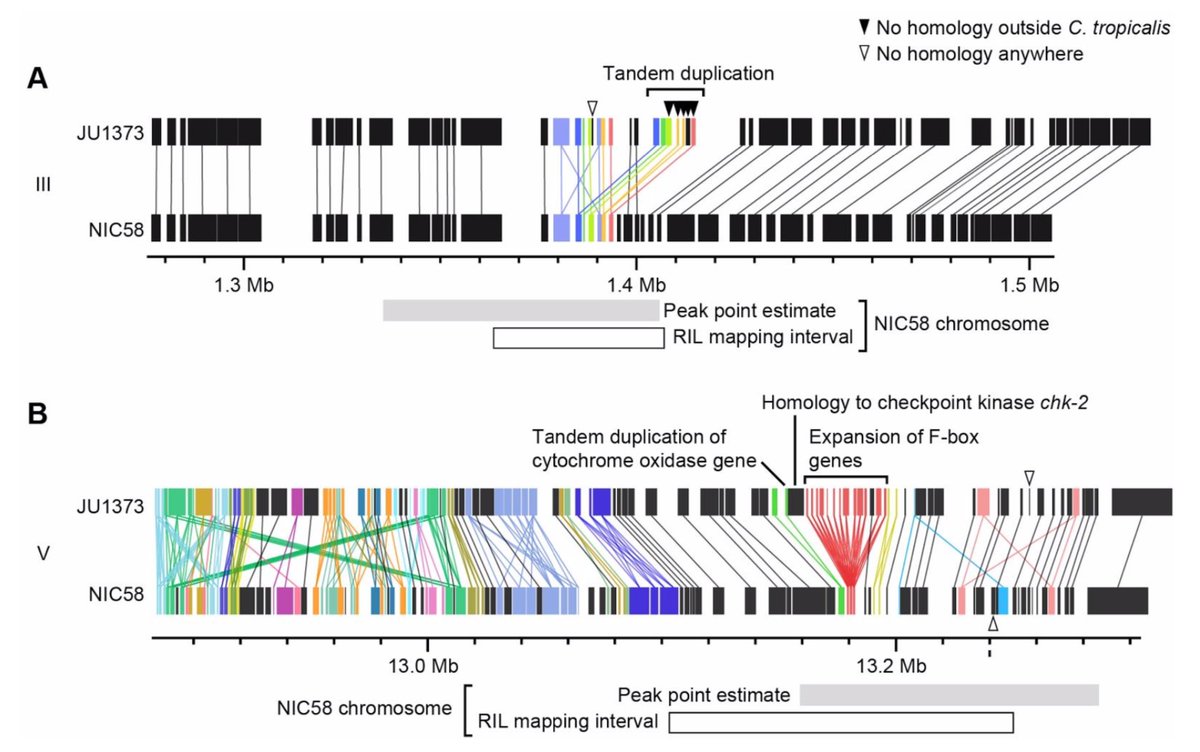
What about outbreeding depression? From surviving RIL genotypes, then a ton of crosses, genotyping, phenotyping and head scratching, we infer the presence of 3 driver elements at 2 divergent loci. As in Burga et al., one of these (on V) is a case of driver antagonism, and all are
consistent with the maternal toxin, zygotic antidote model shown for 2 drivers in elegans by Hannah @wormsrock, @eyalbenda @arburga with @leonidkruglyak. They are widespread, sometimes segregating within populations, and are modified by mitochondrial and nuclear interaction.
We haven't nailed down the genes, but both of the small intervals contain clear candidates in duplicated, or completely novel, genes.
So, we think heterogenous genetic diversity in tropicalis reflects frequent selfing (very low background due to indirect selection on linked sites) and preservation of ancestral diversity by balancing selection of various forms, including local adaptation.
Why are drivers so common in tropicalis? 1 idea is that the high selfing rate renders them neutral, since they are only active in heterozygotes. But could selfing rate also be a consequence of drivers? We think so. This has been seen in hybrid zones, and modelled for CRISPR.
Multiple 'stacked' drivers are difficult to escape.
Work from @weird_worms @asherdcutter et al. shows tropicalis herms are uniquely resistant to the sterilising effects of sperm from dioecious spp., suggesting they have diverged in sperm guidance cues. https://bit.ly/killersperm
Work from @weird_worms @asherdcutter et al. shows tropicalis herms are uniquely resistant to the sterilising effects of sperm from dioecious spp., suggesting they have diverged in sperm guidance cues. https://bit.ly/killersperm
And we see heritable variance in hermaphrodite and male outcrossing probability, and in spontaneous male occurrence; so there's plenty of scope for tinkering with selfing rate. This does not answer "why tropicalis?" and we can only speculate at this point:
maybe this feedback amplified initial differences in Ne among the selfers, maybe tropicalis has evolved faster and/or longer? Thoughts are very welcome! There's much, much more in the preprint.
I can also mention now that the set-up for this work was dicey, but seems to have worked out. @mbeisen laid the groundwork, @MCHammer nailed it down, @Lewis17Stevens capitalized first and well, I thank them all.

 Read on Twitter
Read on Twitter