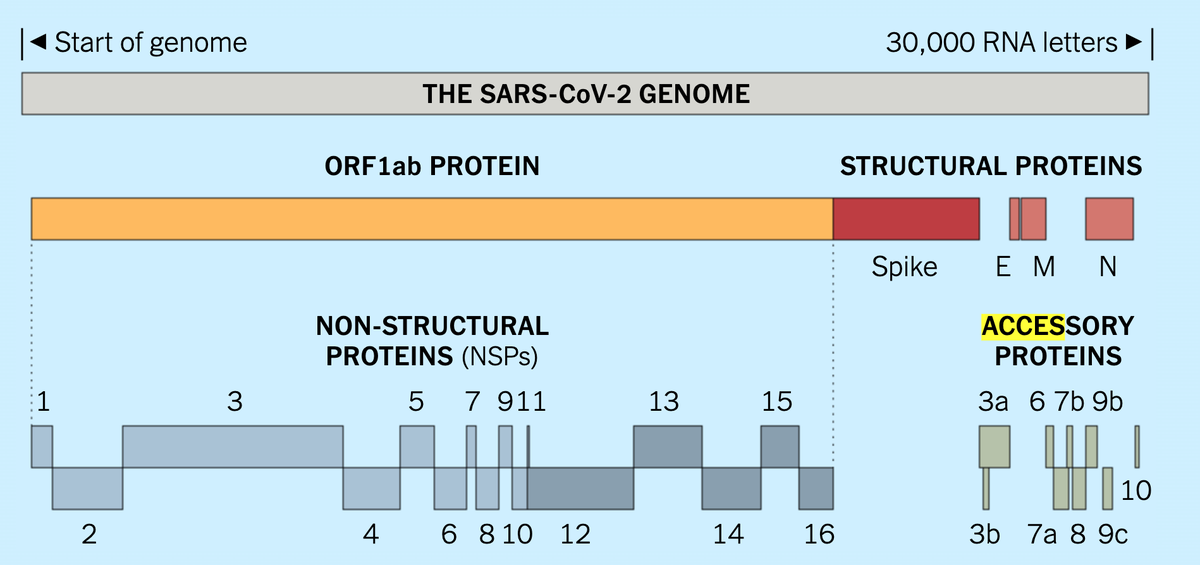
What are all the molecular parts of #SARSCoV2? Getting the answer out of existing databases is harder than you might think. I wrote an article on our work curating a canonical list of proteins for our COVID-19 #KnowledgeGraph https://douroucouli.wordpress.com/2020/08/05/what-is-the-sars-cov-2-molecular-parts-list/ 1/n
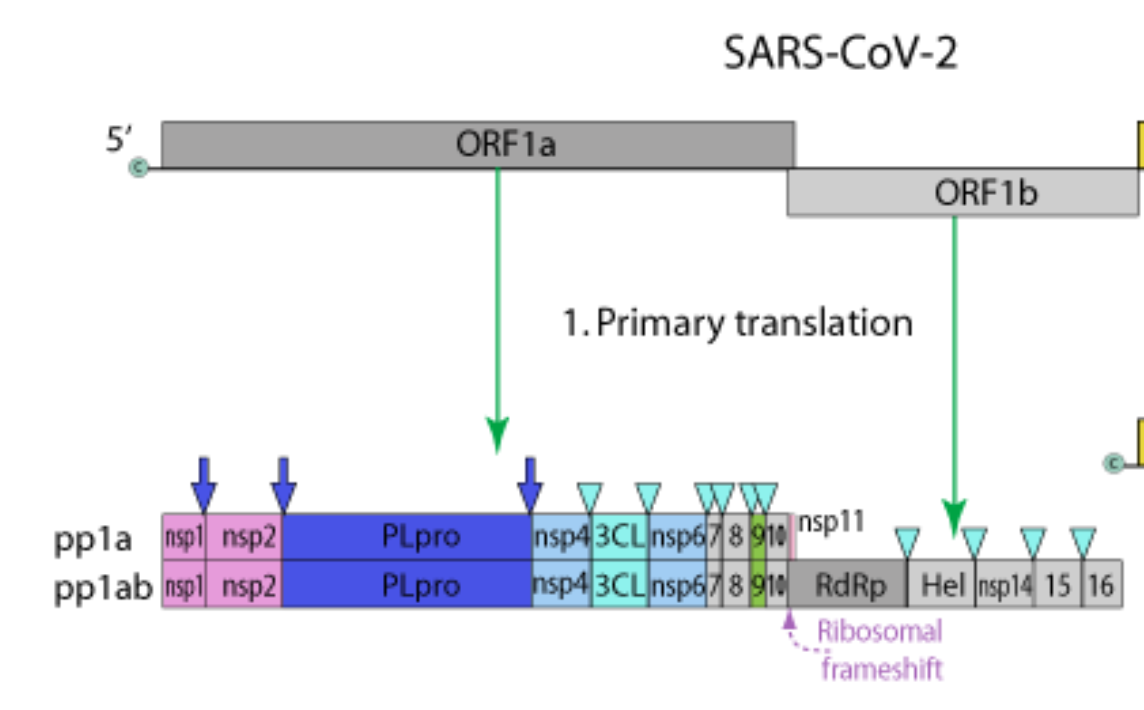
The challenge arises from the existence of viral polyproteins. These are less common in non-viral genomes (but not unheard of - e.g human POMC gene encodes a polyprotein). Each polyprotein is cleaved into multiple discrete functional proteins, as shown below 2/n
Unfortunately these cleaved products (NSPs in SARSCov2) are not always treated as first-class entities in many databases, even though these are the 'parts' we are most interested in our parts list. For example, there is no entry in NCBI Gene, and no accession in UniProt 3/n
Additionally, the presence of two overlapping polyproteins with identical sequences up until the frameshift, results in 'pseudo-duplicates', in which parts lists show two entries, where there should in fact be one (e.g. for nsp1 below). 4/n
Full details of our approach in the post, but working with @news4go @uniprot @ISBSIB @SciBite @intact_project #ProteinOntology curators, @realmarcin produced our canonical parts list, using uniprot accessions + chain IDs https://github.com/Knowledge-Graph-Hub/kg-covid-19/blob/master/curated/ORFs/uniprot_sars-cov-2.gpi 5/n
This is now being used in our KG ( https://github.com/Knowledge-Graph-Hub/kg-covid-19/) and also as a substrate for viral pathway GO-CAM curation, as well as a vocabulary for NLP efforts, e.g. @CovidScholar. File in in GitHub, PRs and issues welcome! 6/6

 Read on Twitter
Read on Twitter