So frustrating for scientists to find that data regarding SARS2 natural origins cannot be accessed. Today, we discovered that the Natural Insertions paper published on June 8 in @CurrentBiology doesn't have its raw data made publicly available. https://twitter.com/shingheizhan/status/1288892478410481664
We are talking about the one paper that describes a bat CoV, RmYN02, that has an "insertion" where the S1/S2 FCS is in SARS2. Note that this PAA site is (1) not a polybasic site, and (2) whether cleavage occurs there remains to be evaluated.
Without the raw data, other experts like @shingheizhan cannot re-assemble the genomes of these novel bat CoVs to check (1) is the sequence correct? (2) is the genome chimeric? (3) does the downstream amplicon sequencing make sense?
Having already endured this mysterious situation with the Guangdong pangolin CoV, @shingheizhan and I are starting to wonder if anyone (authors, journal) even checks that these raw data exist and have been made available by the date (or at least within the week) of publication.
Maybe other scientists can try to find the raw data for RmYN02. Here is the info: China National Microbiological Data Center (project accession number NMDC1001304 and sequence accession numbers: NMDC60013004-01, NMDC60013004-02, and NMDCN0000001-NMDCN0000003). Is login required?
The reason why we are interested to look at the raw data is because the RmYN02 sequences (as well as those of RmYN01) come from a pooled sample = combination of 11 fecal samples from Rhinolophus malayanus collected between May 6 and July 30, 2019.
When the authors checked each of the 11 samples, RmYN02 and RmYN01 were only detected in sample no. 123 collected on June 25, 2019.
To build the genome of RmYN02, the clean reads of pool 39 were assembled. The sequences of the S1/S2 cleavage site and the 1b (RdRp) were verified using amplicons (primers provided, but not the PCR data).
Looking at amplicon data, 1b is sequenced from 12002-19257 with primers F/R1-4. The S1/S2 site was checked with F/R6 from 23070-25439.
We're missing 19258-23069. Could it be that the missing primers F/R5 were intended to check if these 2 fragments (1b and Spike) are connected?
We're missing 19258-23069. Could it be that the missing primers F/R5 were intended to check if these 2 fragments (1b and Spike) are connected?
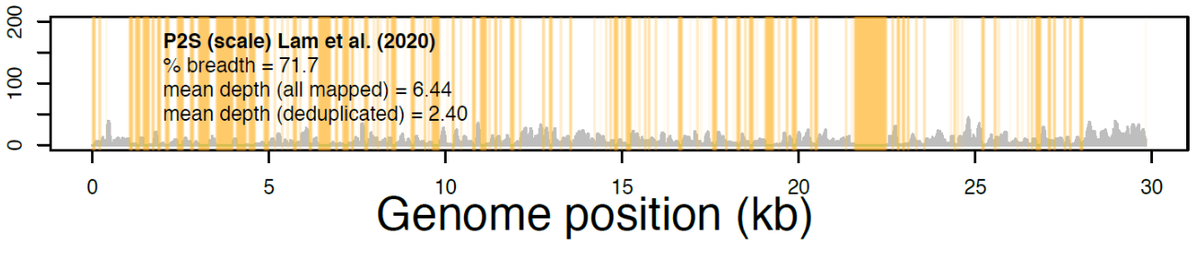
This struck me as a coincidence because that region is also missing from the Lam et al. raw data for the Guangdong pangolin CoV. See the big orange zero read zone between 21-23kb.
Another coincidence? The genomic sequences in the Natural Insertions paper were provided by Professor Wuchun Cao and Professor Yi Guan - both not authors of the Curr Biol paper but authors of the Lam et al. pangolin CoV paper.
In sum, closest relatives to SARS2:
RaTG13 - very slowly revealed origins tied to miners infected by unknown virus, manifesting SARS-like symptoms in 2012
GD pangolin CoV (with the SARS2 RBD) - data missing and confusion among papers
RmYN02 (natural insertion) - data missing
RaTG13 - very slowly revealed origins tied to miners infected by unknown virus, manifesting SARS-like symptoms in 2012
GD pangolin CoV (with the SARS2 RBD) - data missing and confusion among papers
RmYN02 (natural insertion) - data missing

 Read on Twitter
Read on Twitter