On the occasion of the #NAF2020 #RSCPoster session, I’m going to present results from my PhD project “Understanding the structural effect of nucleic acid damage due to metal dysregulation” underneath this tweet ↓ so let’s go #Crystallography #CD 1/29
Before going straight to the point, I will give some context. It is important to know that metal dysregulation in cellular environments has been linked to neurodegenerative diseases such as Alzheimer’s diseases and that it has been shown to lead to oxidative stress. 2/29
Oxidative stress is essentially an imbalance between antioxidant agents and reactive oxygen species (ROS), ROS that can cause extensive damage to cellular components via the Fenton reaction. 3/29
The problem with the Fenton reaction is that it is one of the key pathways to produce hydroxyl radicals by reaction between hydrogen peroxide and transition metal ions and that it can cause oxidative damage to multiple bases in Nucleic Acids. 4/29 https://pubs.acs.org/doi/pdf/10.1021/acs.joc.5b00689
In their paper, Hofer et al. has linked an increase in oxidative damage (with an increase of the major oxidation product 8-oxo-7,8-dihydroguanine, or oxoGua) with a high intracellular neuronal iron levels In Alzheimer’s diseases 5/29 https://www.sciencedirect.com/science/article/pii/S0946672X16300967
The aim of my PhD project is to monitor the effects of these oxidative damage as a function of structure, and for that I am using both X-ray crystallography and Circular Dichroism (CD). 6/29
Now to try and model this reaction on the atomic scale, I crystallise nucleic acids with various transition metals to obtain structural data. The endgame here is to attempt in-crystal Fenton reaction and identify damage hotspots in each structure. 7/29
But before I get there, I need to find nucleic acid crystals that will allow me to soak in transition metals (whether biological relevant i.e. iron, copper or models i.e. cobalt nickel) with the hope that I’ll be able to soak in Fenton reagents in the future. 8/29
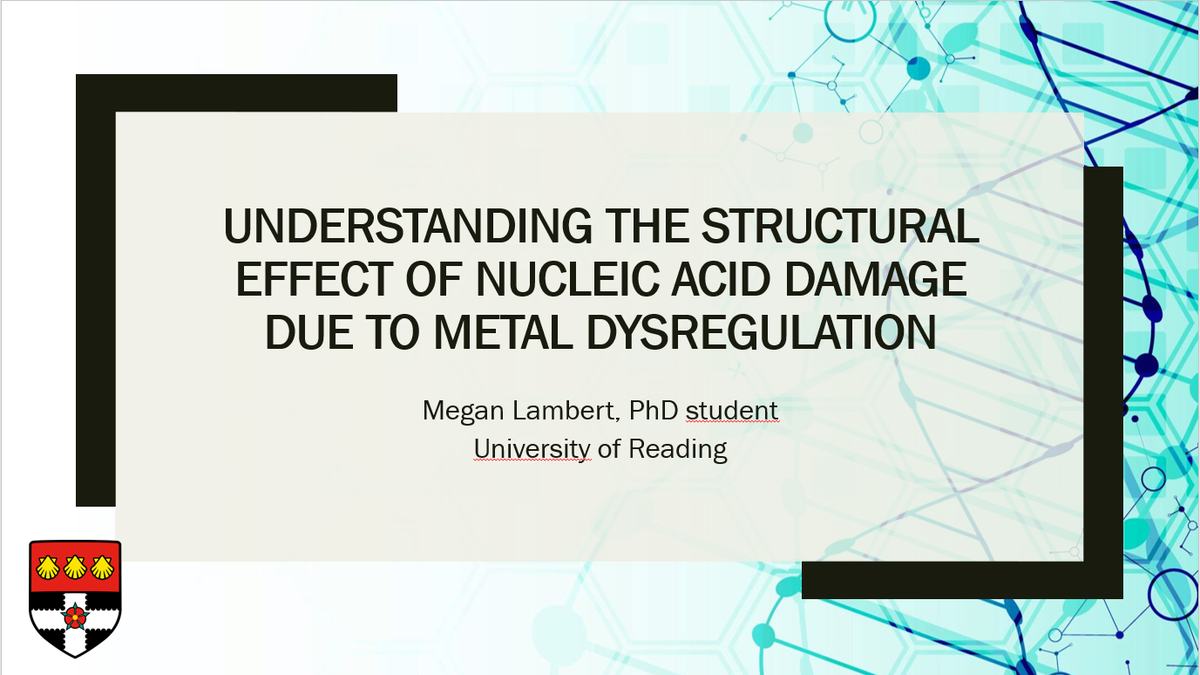
Crystals of the DNA sequence d(GCATGCT) allow me to do that. The crystals were grown at 291 K using sitting-drop vapor diffusion. 9/29
In this method, drops containing nucleic acids and crystallisation conditions are allowed to equilibrate with a larger reservoir. With the right conditions, crystal growth occurs. 10/29
For d(GCATGCT), the drops contained 2μL of 1mM DNA, 1μL of 10% (v/v) MPD, 1μL of 40mM sodium cacodylate trihydrate pH 5.7, 1μL of 80mM hexamine cobalt (III), 2μL of 30mM sodium chloride and 2μL of 800mM potassium chloride. 11/29
In this case, the drops were equilibrated against 500μl of 35% (v/v) MPD and the crystals grew within 2 months. The crystals were then soaked in 25mM copper sulfate for 24 hours. 12/29
X-ray diffraction data were collected on an in-house X-Ray source at the University of Reading and the data processed using CrystallisPro. The structures were solved using molecular replacement and refined using the Phenix software suite. 13/29
Results: Cu (II) ions interact with the DNA by coordinating to the N7 position of the guanines but appear to bind only to guanines in the first positions. However, all copper ions are bound to guanines available on the outside of the structure. 14/29
Now that I identified possible reaction centres, I can now attempt to induce in-crystal Fenton reactions, to examine the structural consequences of damage of neighbour bases at atomic resolution. 15/29
Of course, X-ray crystallography is not the only method to study nucleic acid structure, it is also possible to use Circular Dichroism (CD). 16/29
CD is a form of light absorption spectroscopy that measures the difference in absorbance of right- and left-circularly polarized light by a substance. It is sensitive to the conformational states of nucleic acids and allows to study different nucleic acid structures. 17/29
It is now easy to associate a CD spectrum to structures thanks to the work of many groups through the years. Some papers like the one published by Kypr et al. review the most common nucleic acid structures. 18/29 https://academic.oup.com/nar/article/37/6/1713/1030465
For this, I focused on G-quadruplexes, four-stranded quadruplexes that can form in G-rich sequences and which are found in the telomeric and gene promoter regions of the genome. As an example, here’s the crystal structure of the human telomeric G-quadruplex (PDB ID: 1KF1) 19/29
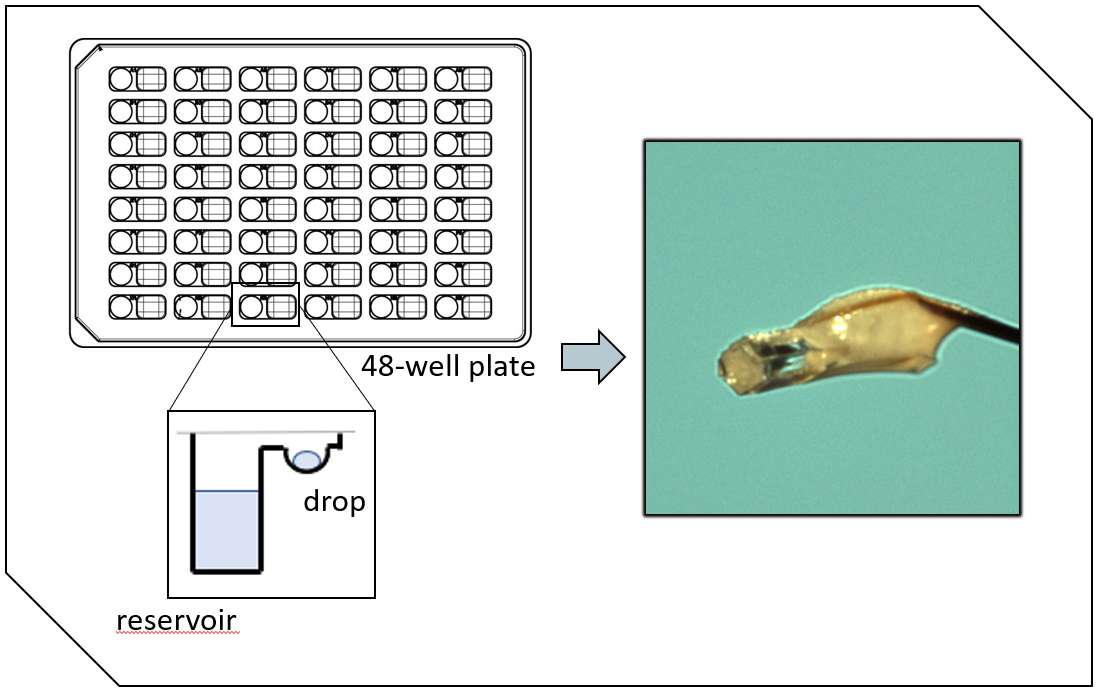
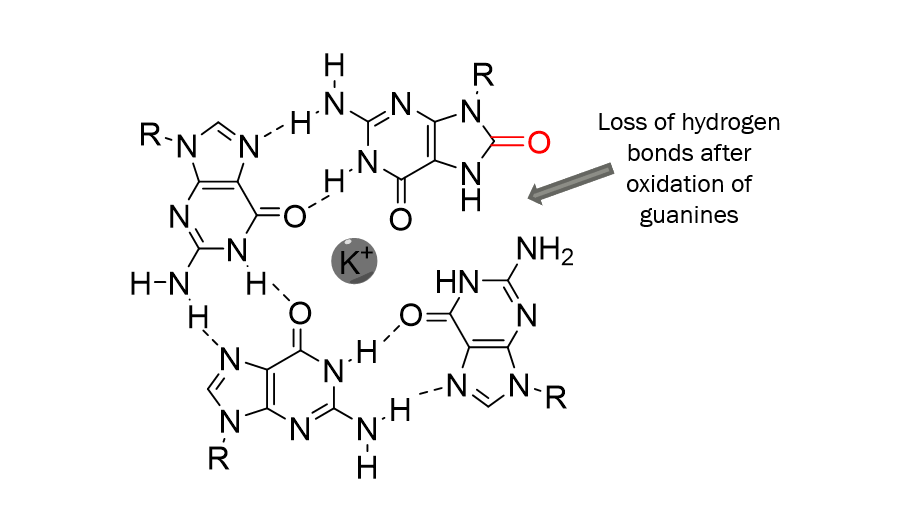
In these structures, four guanine bases can associate through what is called Hoogsteen hydrogen bonding to form a square planar structure called a guanine tetrad, stabilised by a cation, usually potassium or sodium. 20/29
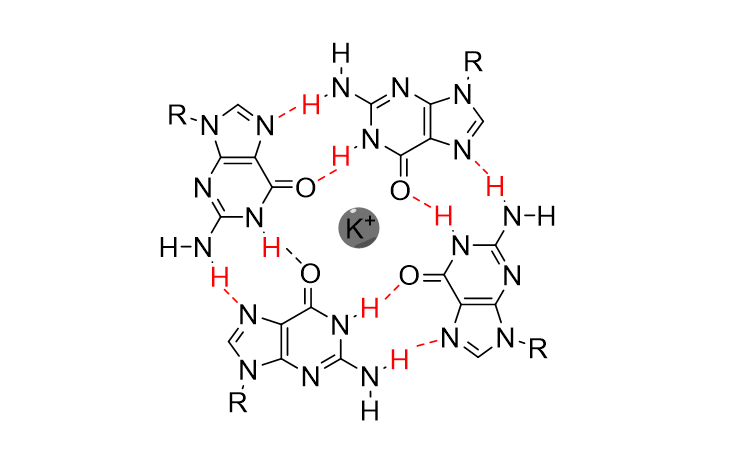
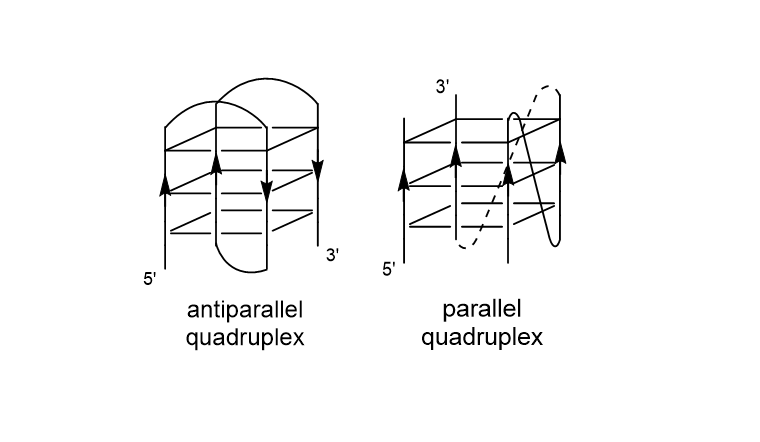
And these tetrads can then stack on top of each other to form the 3D structure. Now, depending on how the guanine bases are arranged, a quadruplex can adopt several topologies: parallel, antiparallel or a mix of both. 21/29
What I tried to do was to see what happens to the structure when a G-quadruplex forming sequence is present in an oxidative stress environment. Here, in the presence of Fenton reagents (hydrogen peroxide and copper (II) ions). 22/29
What I expected was a destabilisation of the whole structure explained by the loss of hydrogen bonds after oxidation of guanines in oxoguanines. 23/29
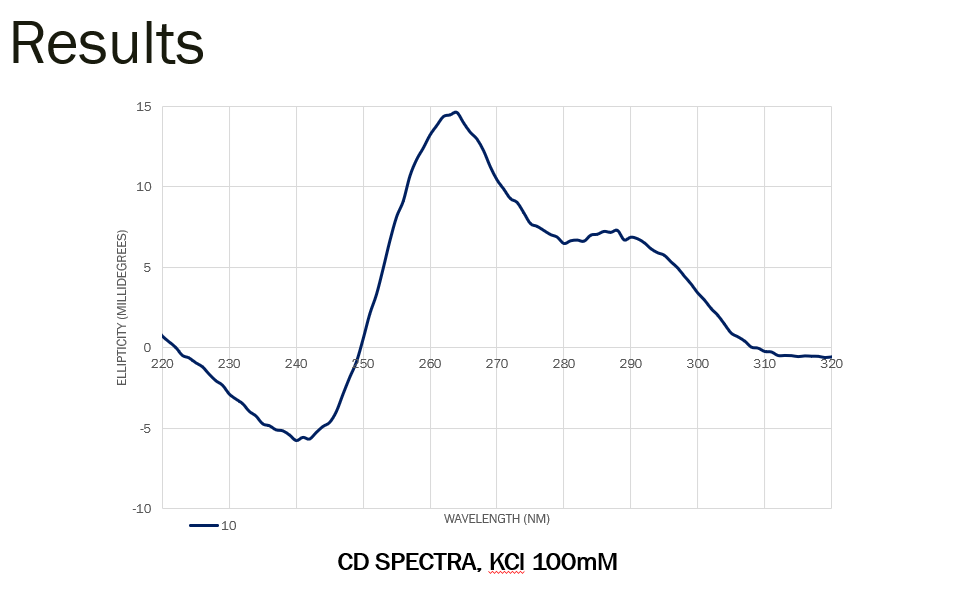
I used a unimolecular G-quadruplex forming sequence and CD spectra in presence of potassium showed two positive peaks at 260nm and 290nm and a negative peak at 240nm, which suggests a mix of antiparallel and parallel strands. 24/29
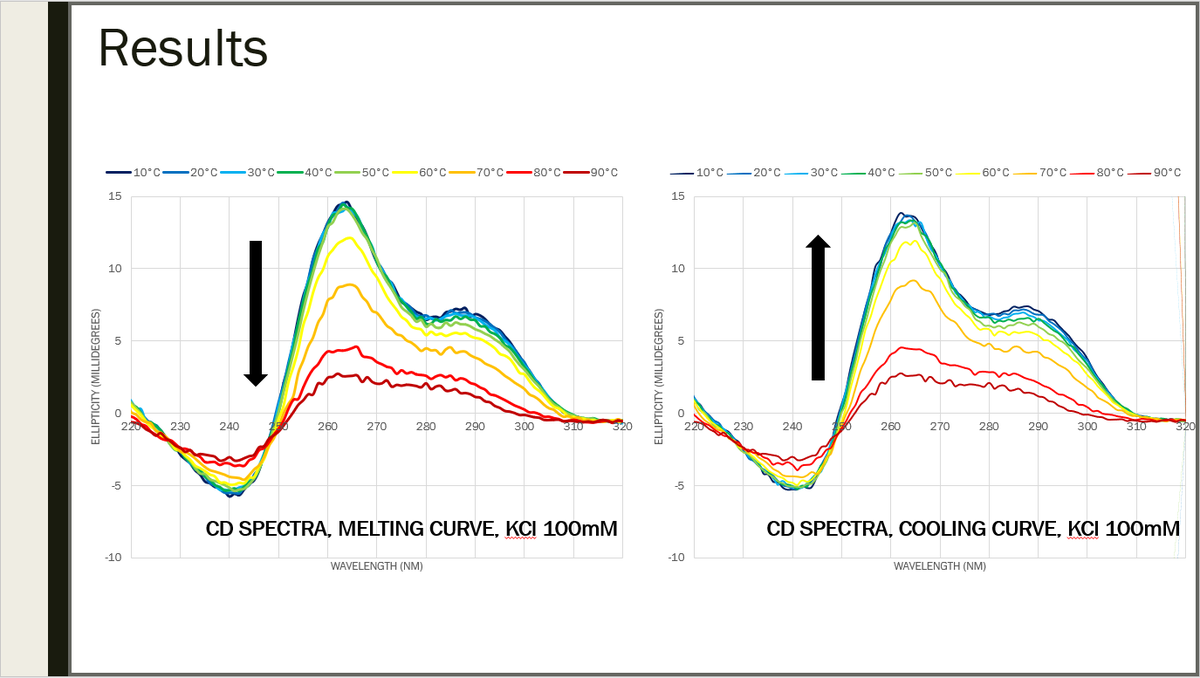
Both melting and cooling were also carried out to check the structure folds and unfold correctly. 25/29
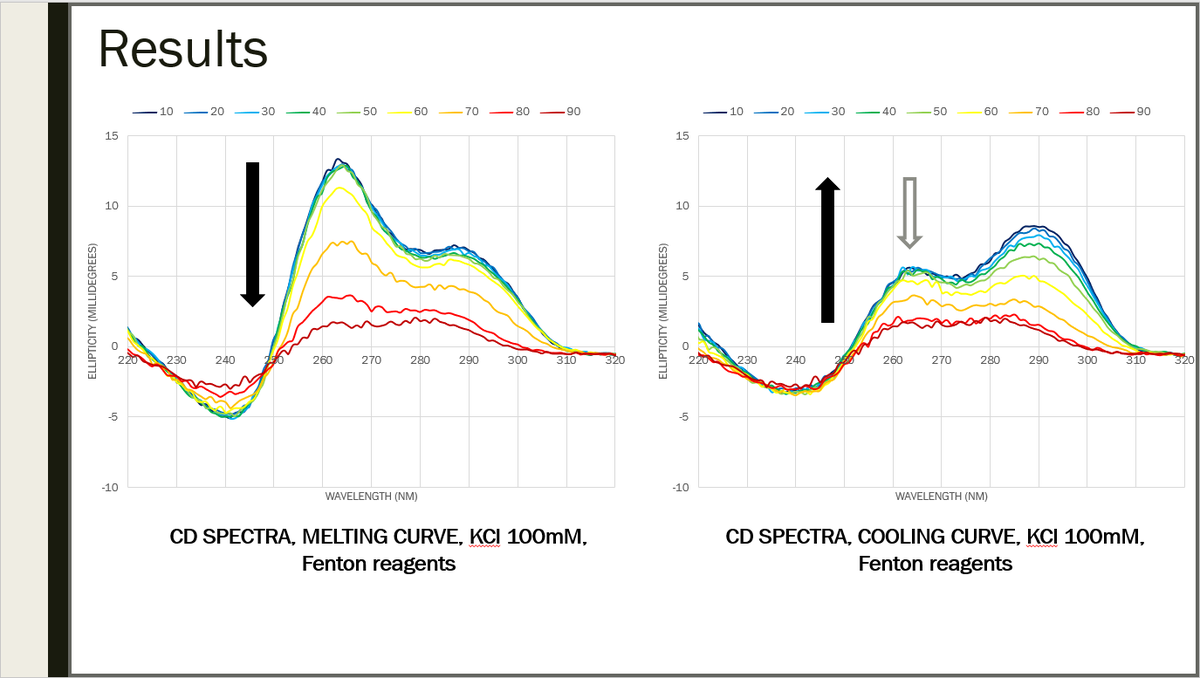
Results after Fenton reagents were added in the solution (CuCl2 200 uM, H2O2 1.5mM, citric acid 1 mM): a loss in the signal at 264nm has been observed after cooling, meaning that damaging this DNA sequence with Fenton reagents prevents it from refolding in the same way 26/29
Of course, this could be explained by either oxidation of guanines or by strand breaks, so to test these hypothesis, native gel electrophoresis will be run with DNA treated with Fenton reagents in the same conditions. 27/29
In the future, I will investigate this structural change and try to figure out if it is linked to the extra G tracks. I will also try reproducing these results with different topologies. 28/29

 Read on Twitter
Read on Twitter