Proud to share our latest work on MHC imputation! We constructed a multi-ethnic high resolution MHC imputation reference panel with more than 21,000 individuals from five diverse populations. Here are some highlights from our study: https://medrxiv.org/cgi/content/short/2020.07.16.20155606v1
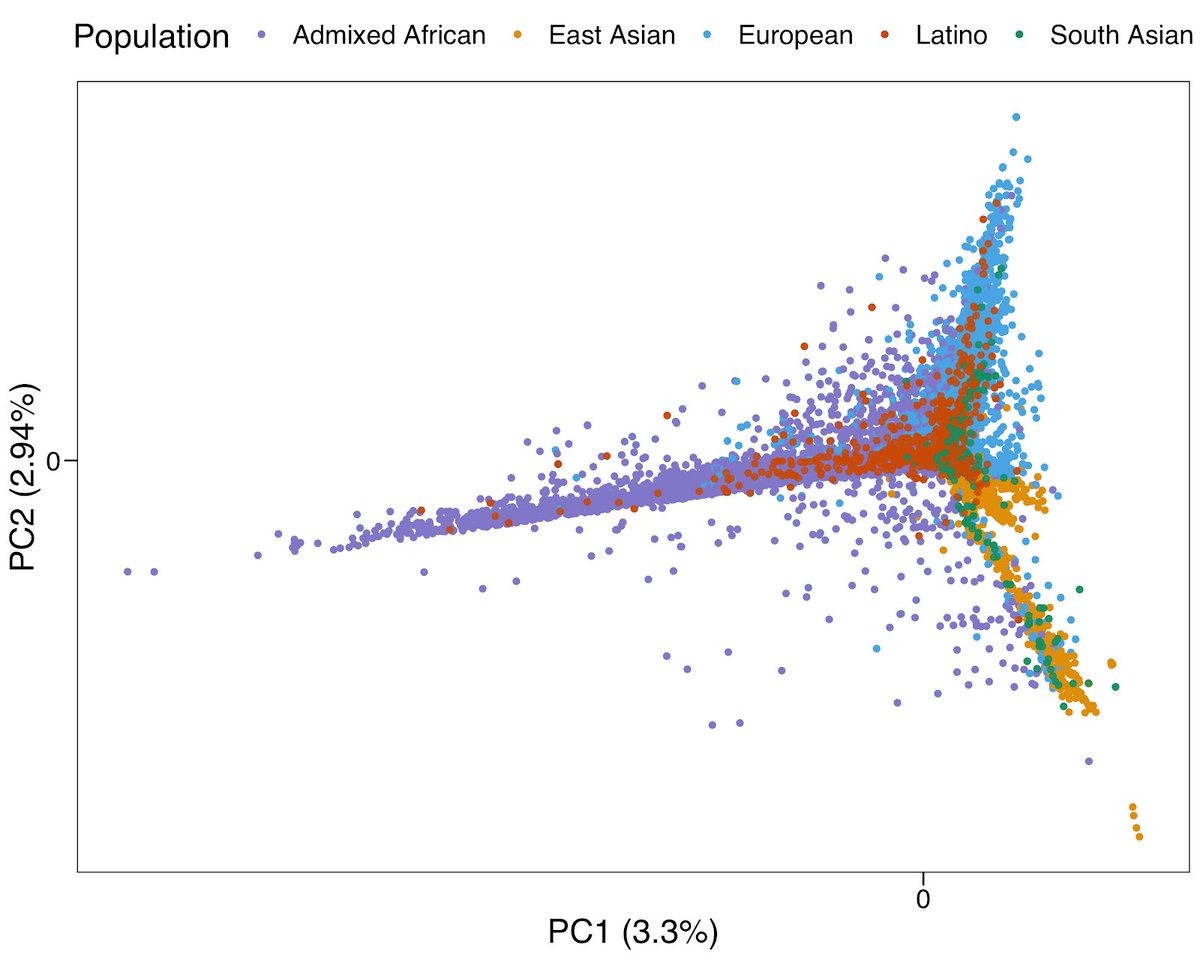
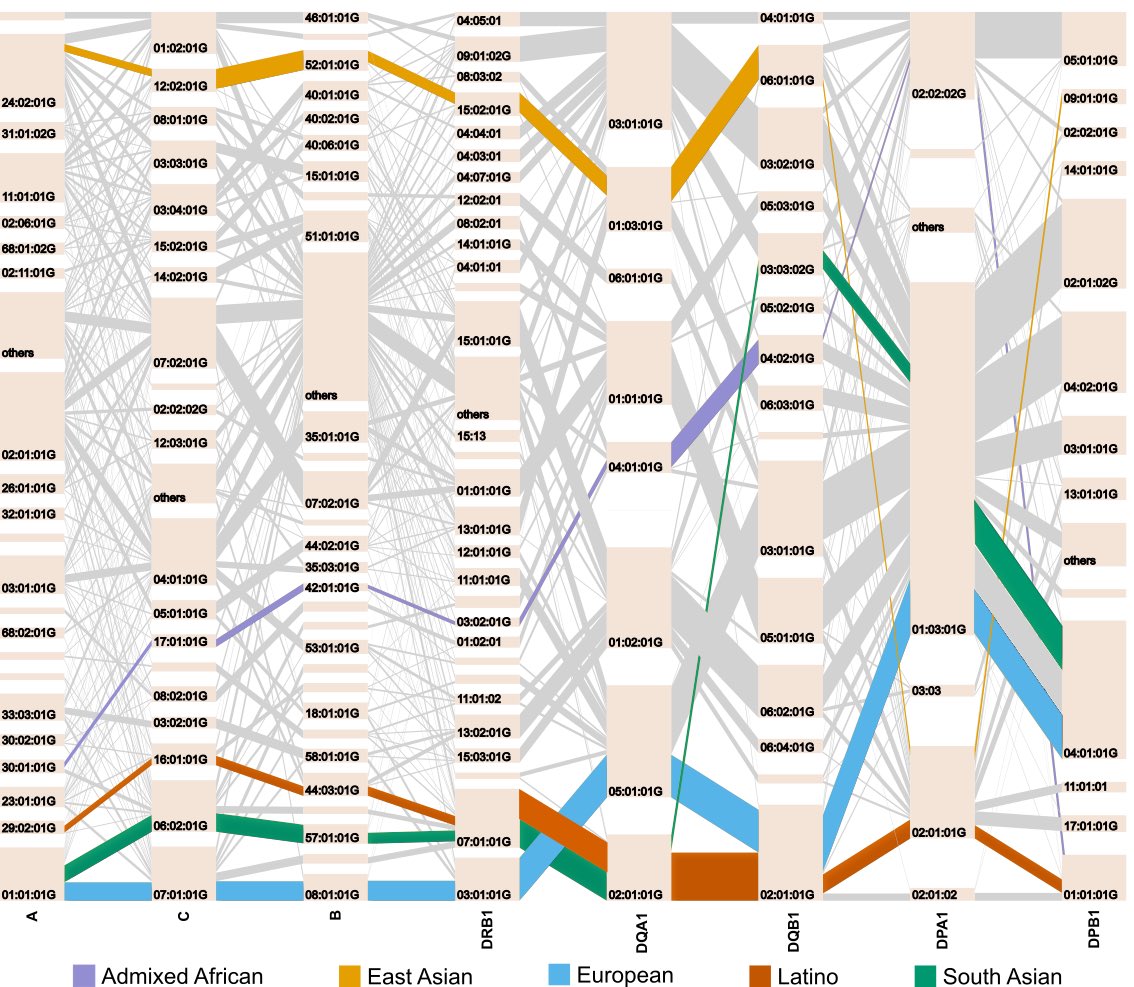
2/6 we observed unique long-range HLA haplotypes within each population group. This highly population-specific feature implies multi-ethnic fine-mapping studies can greatly improve the power for defining causal variations in the MHC region.
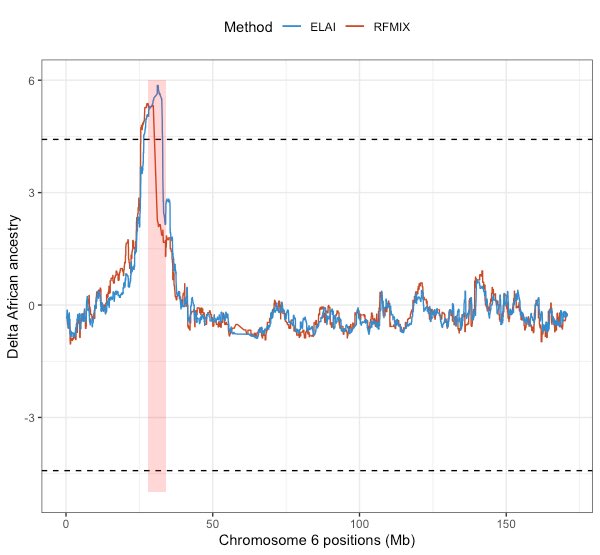
3/6 Using the admixed WGS samples included in the panel, we assessed evidence of selection within HLA, and observed MHC-specific excess of African ancestry in Latinos.
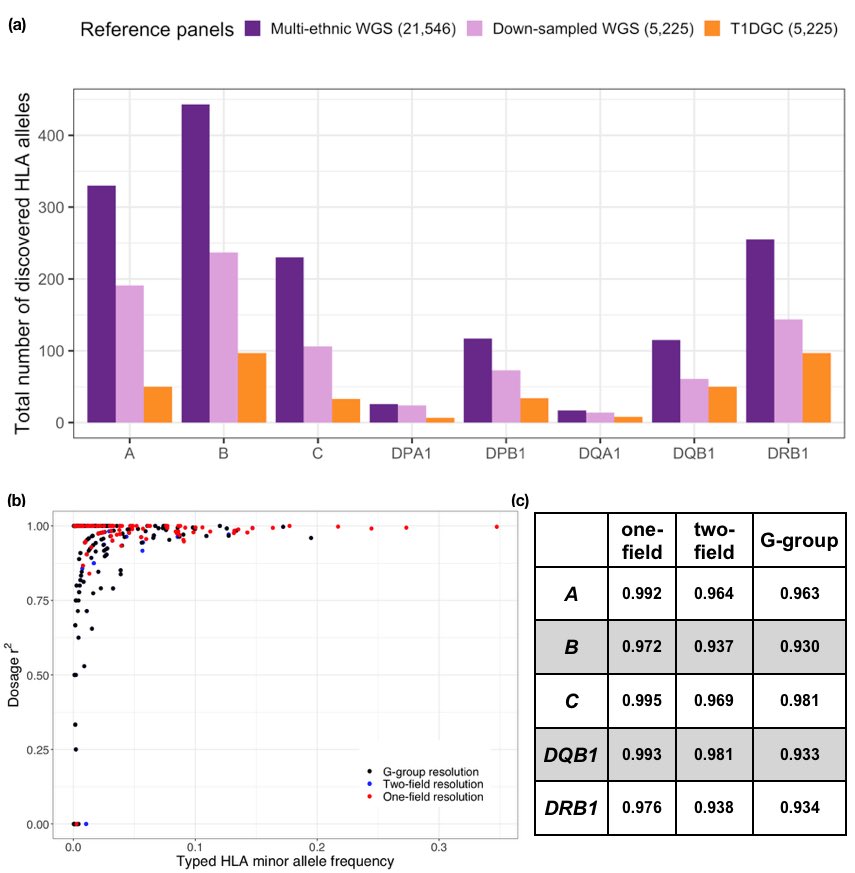
4/6 We showed that the constructed multi-ethnic panel can be used for highly accurate imputation in multiple populations.
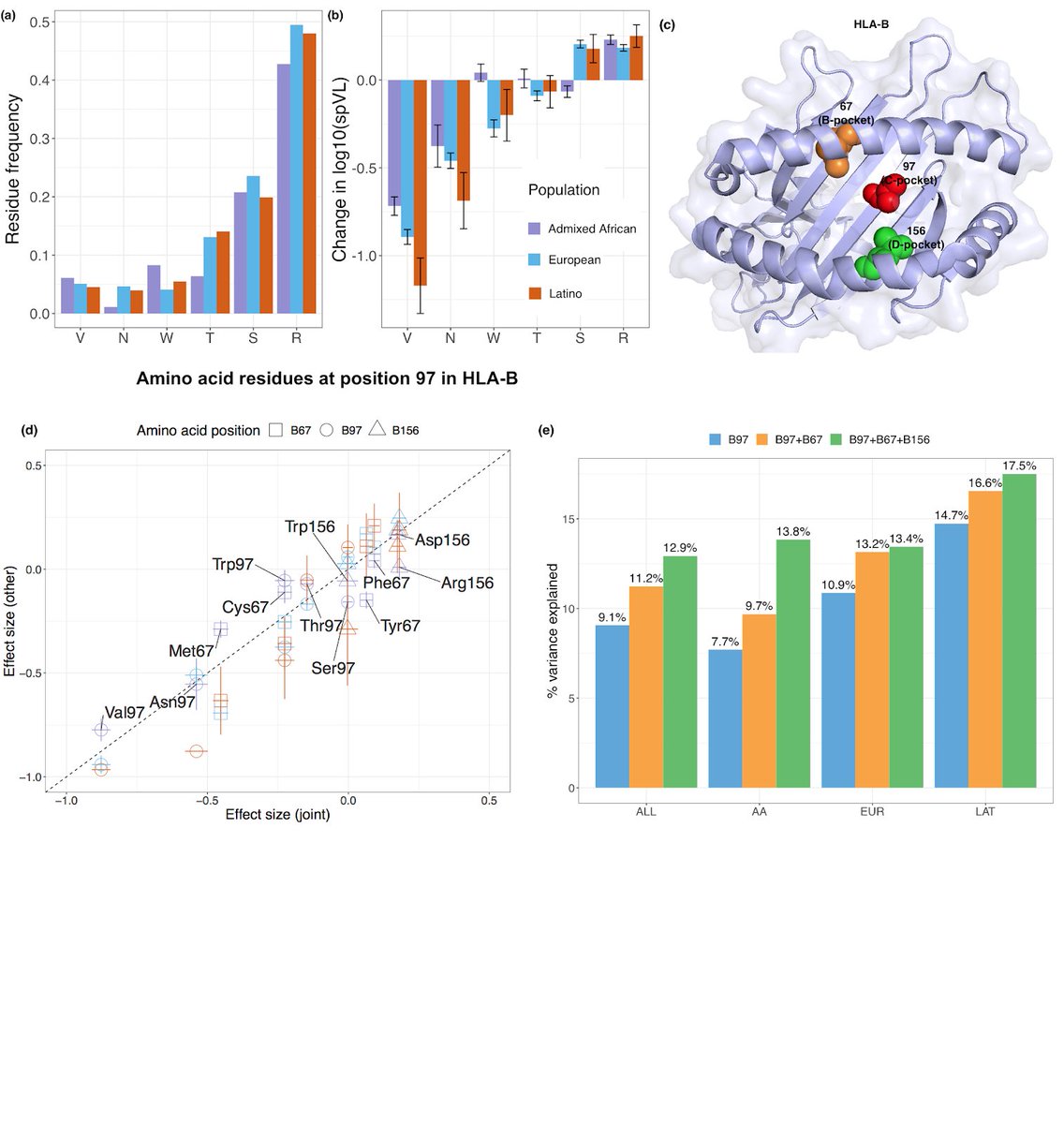
5/6 In a multI-ethnic HIV-1 viral load fine-mapping study, we pinpointed the MHC association to three amino acid positions (97, 67 and 156) marking three consecutive pockets (C, B and D) within the HLA-B peptide binding groove, and obviating effects of previously reported assocs.
6/6. We built a new HLA-focused pipeline that can handle HLA reference panel construction (MakeReference), HLA imputation (SNP2HLA), and HLA association (HLAassoc). https://github.com/immunogenomics/HLA-TAPAS
This work won’t happen without help and support from my amazing colleagues and mentor. @masakanai, @sarahlxy619, @EskoTonu, @okada_yukinori, @fakepaulmclaren and @soumya_boston and many others that are not on twitter!
Special thanks also goes to the amazing WGS efforts from @nih_nhlbi #TOPMed, Japan Biological Informatics Consortium , Biobank Japan, @ESTbiobank , @1000genomes. This whole project won’t happen without the valuable data that they generated!

 Read on Twitter
Read on Twitter