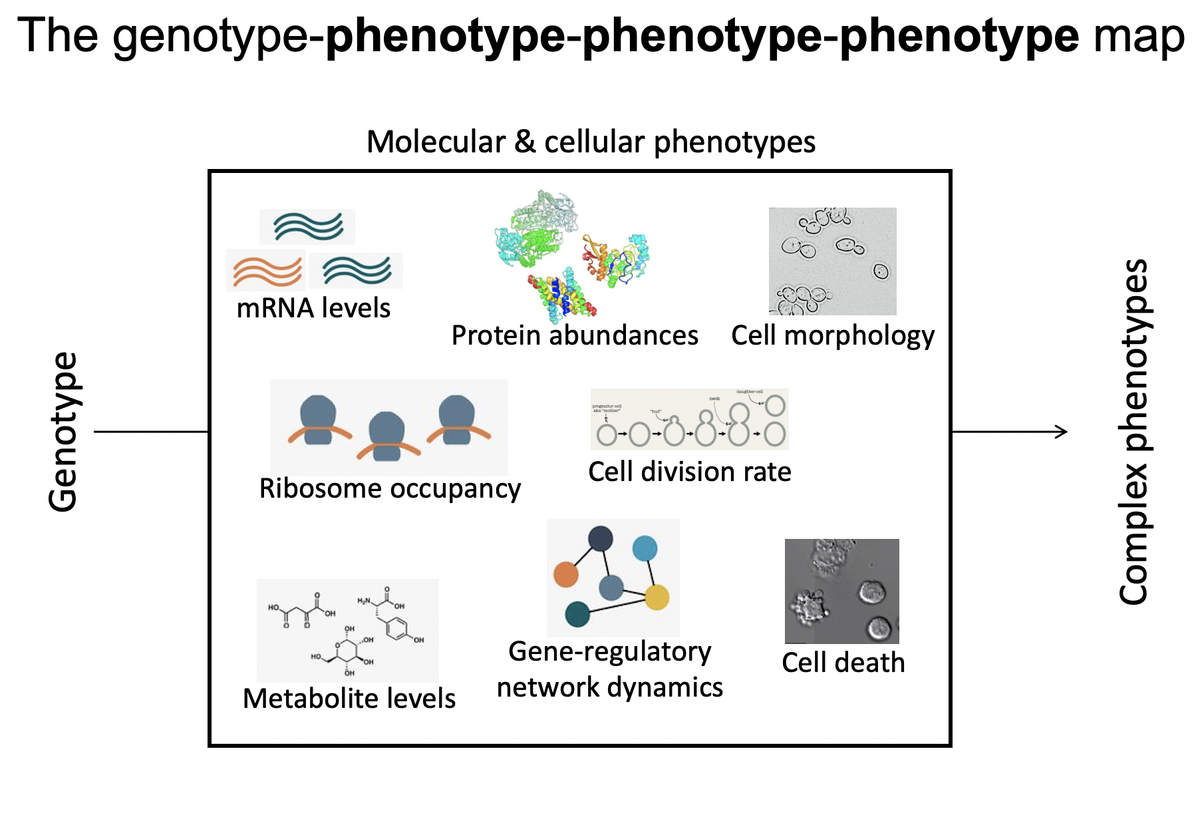
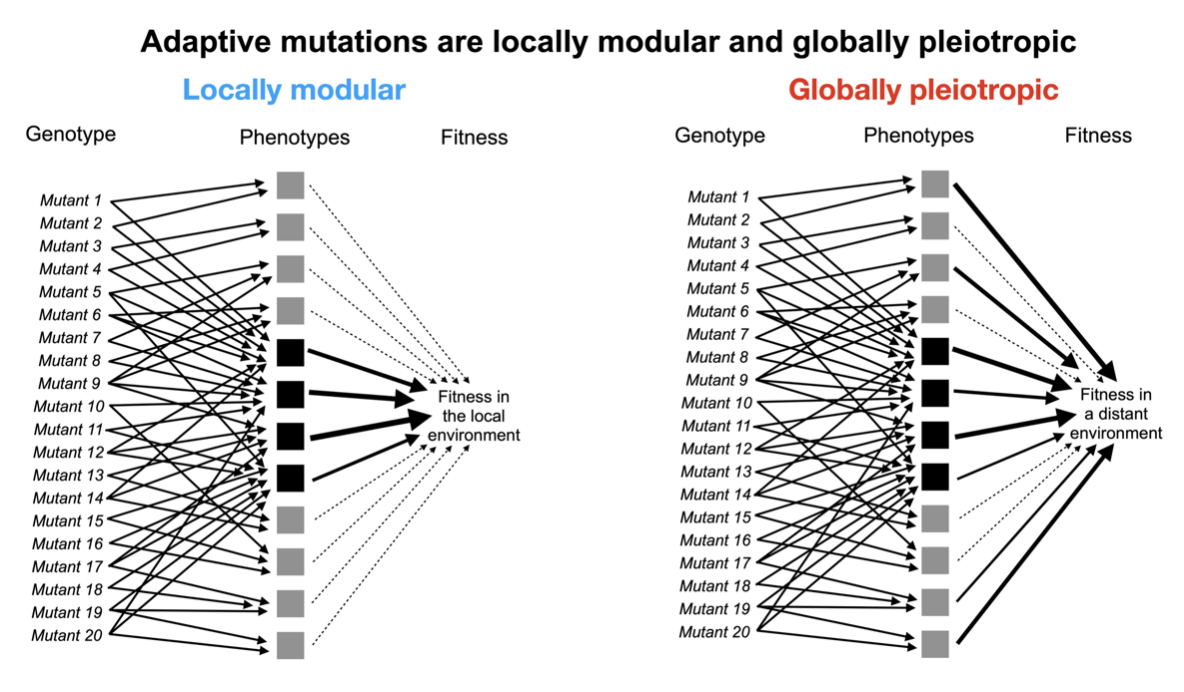
How do you count the # of independent traits a mutation affects (i.e. its degree of pleiotropy)? A yeast cell has thousands of measurable features (e.g. gene expression levels) that are related in complex ways and that influence higher-order traits like cell shape & growth 1/16
It is difficult to imagine how adaptation happens if mutations that make beneficial changes to one trait have collateral effects on thousands of others. If mutations affect many traits, it is also difficult to map genotype to phenotype. Is the G-P map tractable? 2/16
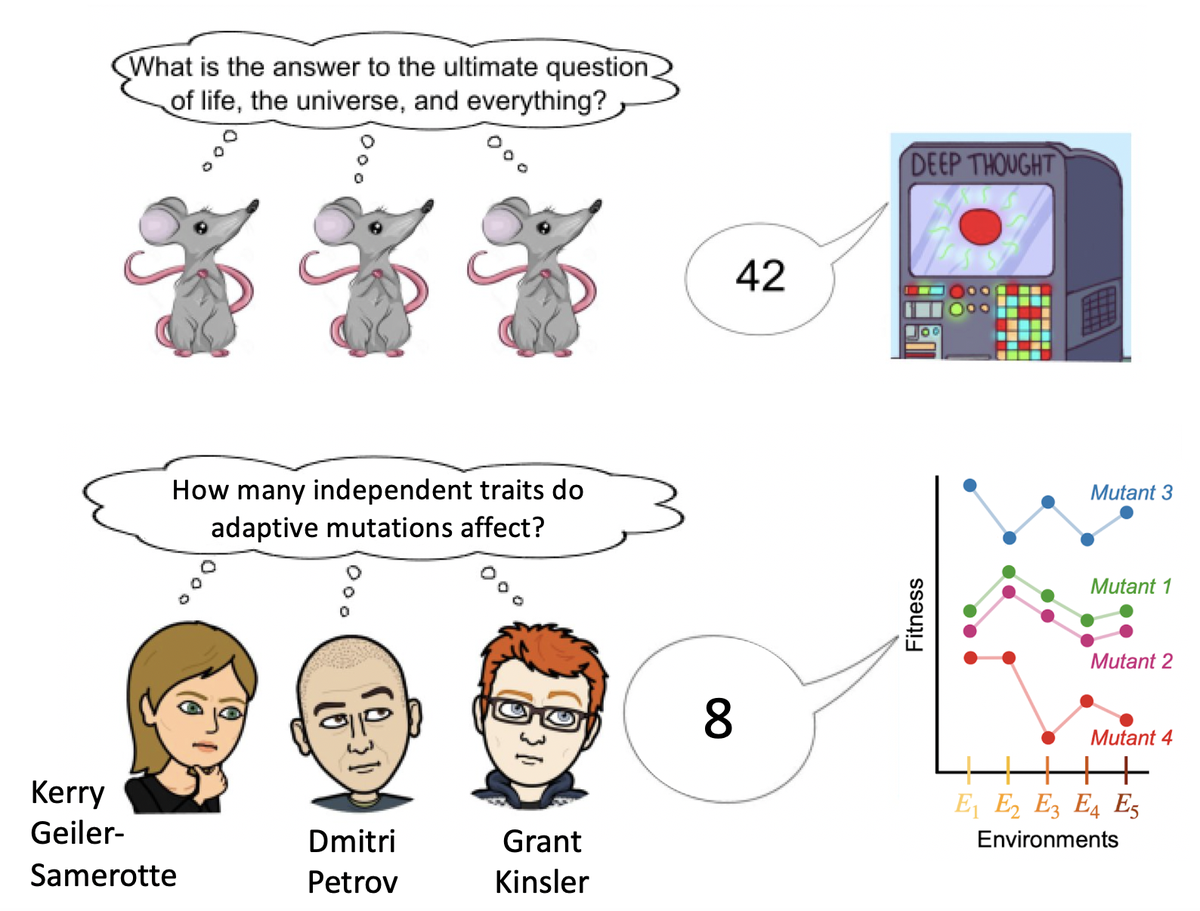
In new work with @PetrovADmitri and @GrantKinsler we present a method to count traits that affect fitness. We find that adaptive mutations affect only a few phenotypes that contribute to fitness in the environment where the mutations originated. 3/16 https://www.biorxiv.org/content/10.1101/2020.06.25.172197v2
This means that the genotype-phenotype-fitness map of adaptation is indeed tractable. We demonstrate this by making accurate fitness predictions for unstudied adaptive mutants (e.g. point mutations in different genes & pathways) in unstudied environments. 4/16
So how does our new method work? I loved being a part of this project because it required the coming together of very different insights and expertise. Also, we had a lot of fun thinking about this stuff. 5/16
My interest in pleiotropy stems from work w/ Mark Siegal. We found the correlation between traits (their degree of independence) changes across contexts like environment & genetic background. How do you map genotype to phenotype when the map changes? 6/16 https://www.biorxiv.org/content/10.1101/700716v2
But @PetrovADmitri @GrantKinsler and I turned this context-dependence problem on its head. We recognized that the fitness impact of each phenotype also changes with context, e.g. a mutation that affects glucose storage might matter more when glucose is scarce 7/16
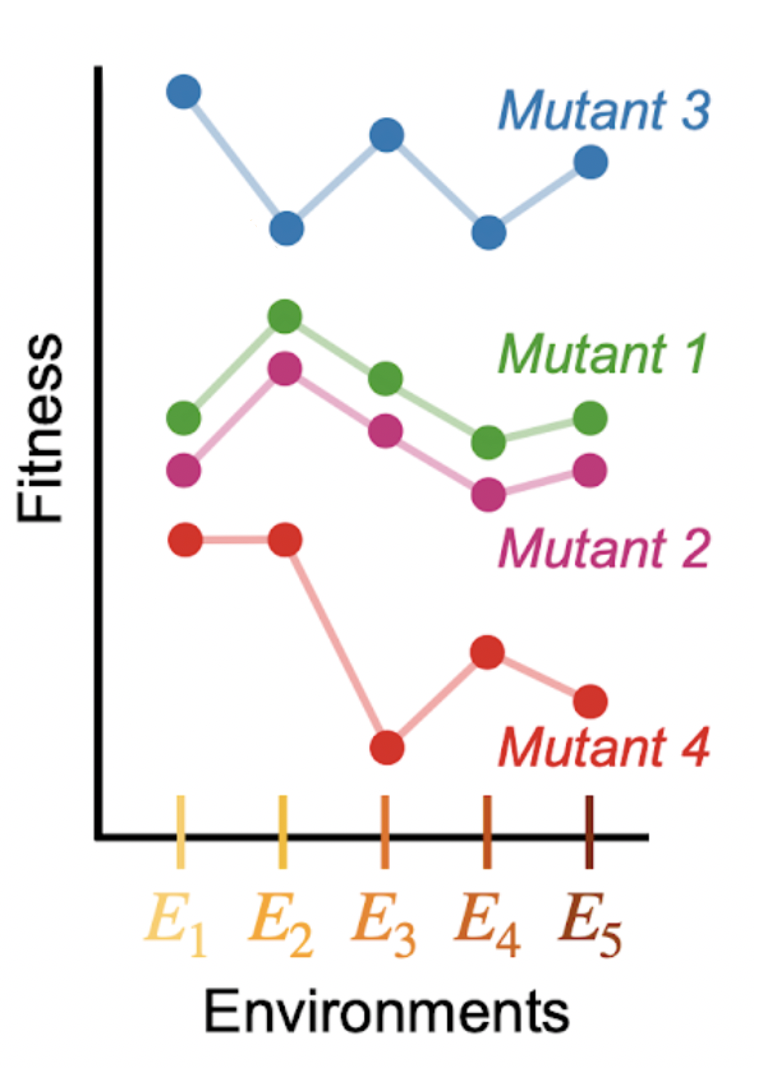
By comparing how the fitness of adaptive mutants changes across many contexts (environments), we can infer which mutants influence similar phenotypes (mutants 1 and 2) and decompose how many phenotypes contribute to fitness overall 8/16
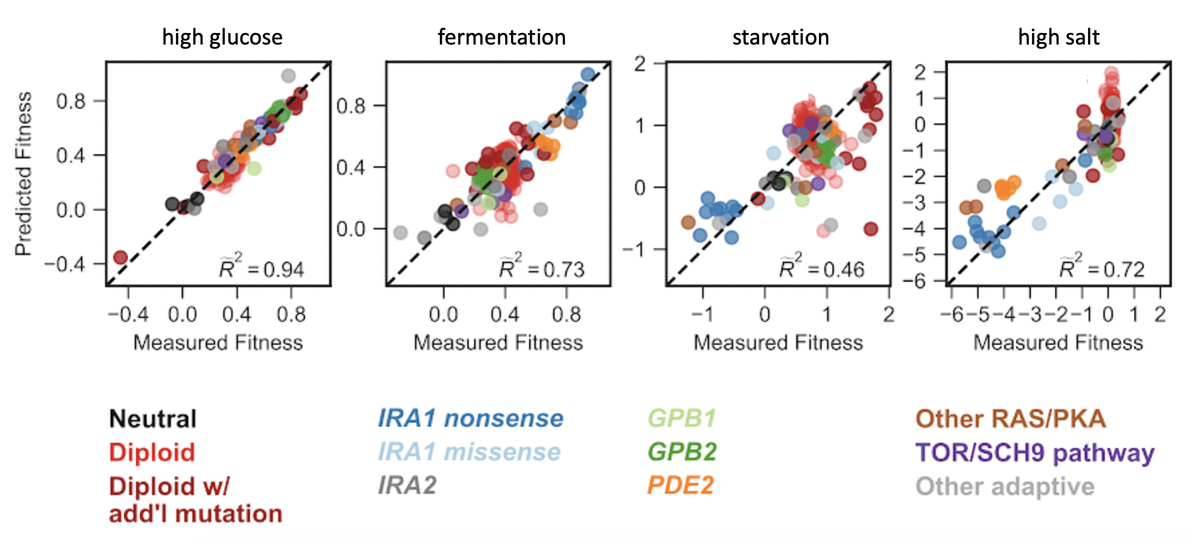
We studied 292 mutants that improved yeast fitness under glucose limitation. Because both the phenotypic and fitness impacts of mutation can change with context, we initially focused on environments that are similar to the one where adaptive mutants evolved. 9/16
Teasing out differences in fitness across subtly different environments is tough! We went crazy reducing sources of noise in what turned into an all-hands-on-deck situation in the Petrov lab. @mmascar @liyuping927 @_ellie_cat We tweeted about these challenges #1BigBatch. 10/16
We captured most fitness variation of 292 mutants across 25 subtly different environments in a genotype-phenotype-fitness model with just a few phenotypic components. These diverse mutants affect similar phenotypes that boost fitness in the environment where they evolved 11/16
But bigger changes to the environment reveal additional phenotypic effects that now matter to fitness (e.g. 3 phenotypic components that explain 1.1% of fitness variation across subtly different environments explain 45% of fitness variation in a starvation environment). 12/16
In sum at the level of fitness adaptive mutations are locally modular, affecting few phenotypes that matter to fitness in the environment where they evolved. Yet they are globally pleiotropic in that they affect additional phenotypes that impact fitness in new environments 13/16
This finding solves the conundrum of how adaptation is possible if mutations affect many traits: Not all of those traits contribute equally to fitness. For another summary of this finding, check out this awesome thread by co-author @GrantKinsler 14/16 https://twitter.com/GrantKinsler/status/1280623025134551040?s=20
This way of studying genotype-phenotype-fitness maps embraces their context dependence. Quantifying how the impacts of mutation change across contexts yields insights about the map’s architecture (the degree of pleiotropy) and improves fitness predictions for new mutations 15/16
Quantifying how the impacts of mutation change with context & understanding the mechanistic basis of these changes is the focus of my new lab @MechofEvolution @asuSOLS. If this interests you check out our review paper @YE_lab @GBilolikar. Or join us! 16/16 https://doi.org/10.1016/j.gde.2019.08.003

 Read on Twitter
Read on Twitter