PAPER ALERT
PAPER ALERT
Introducing DiffNets, a deep learning approach to identify structural features responsible for the biochemical differences between protein variants. This thread is about the work and our plans to apply it to F@h-generated #COVID19 data! https://www.biorxiv.org/content/10.1101/2020.07.01.182725v1
A protein adopts millions of structural poses. Often, protein mutations only subtly alter this "landscape", but dramatically modify a protein's biochemical properties. Its difficult to identify underlying structural features that explain biochemical differences between variants.
For example, previous work in @drGregBowman lab identified structural features that modulate stability across four protein variants, which affects the protein’s ability to degrade an antibiotic drug, cefotaxime. This analysis took months, but can be done in *days* with DiffNets.
That work can be found here: https://pubs.acs.org/doi/full/10.1021/acscentsci.7b00465
DiffNets perform a dimensionality reduction on simulation data that separates protein structures based on their biochemical properties. It is easy to use the resulting low-dimensional projection of data (i.e. map) to identify structural features that distinguish variants.
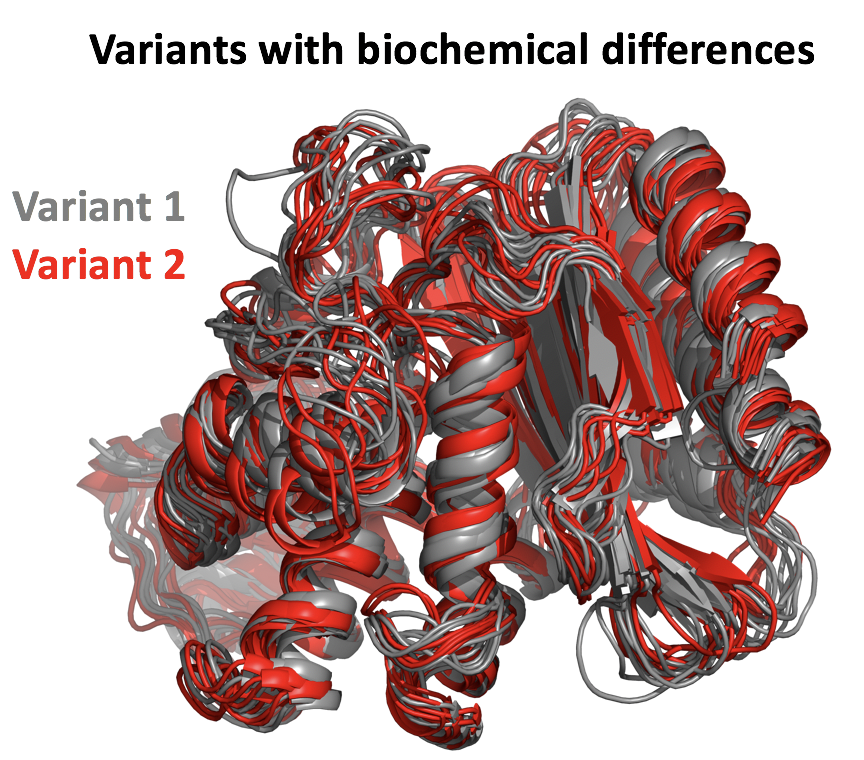
We tested DiffNets on beta-lactamase variants with different stabilities as a proof of concept. DiffNets identify structural features that increase stability of variants, including an extremely subtle helix compaction on the order of < 0.5 Angstroms.
We will use DiffNets to find differences between homologous coronavirus proteins to build on our recent #COVID19 work. We expect DiffNets will aid in identifying mechanistic differences that explain differences in lethality + transmission between viruses. https://www.biorxiv.org/content/10.1101/2020.06.27.175430v1
The code is freely available at https://github.com/bowman-lab/diffnets -- shoutout to @mickdub29 @mizimmer90 and @drGregBowman for this exciting work!

 Read on Twitter
Read on Twitter