Pleased to report ConFiG – Contextual Fibre Growth, our white matter digital phantom generator for developing & validating μ-structure imaging techniques, just appeared @Neuroimage_EiC ( https://doi.org/10.1016/j.neuroimage.2020.117107), joint work with @fishpiechicken, @MarcoPalombo3, @garyhuizhang (1/8)
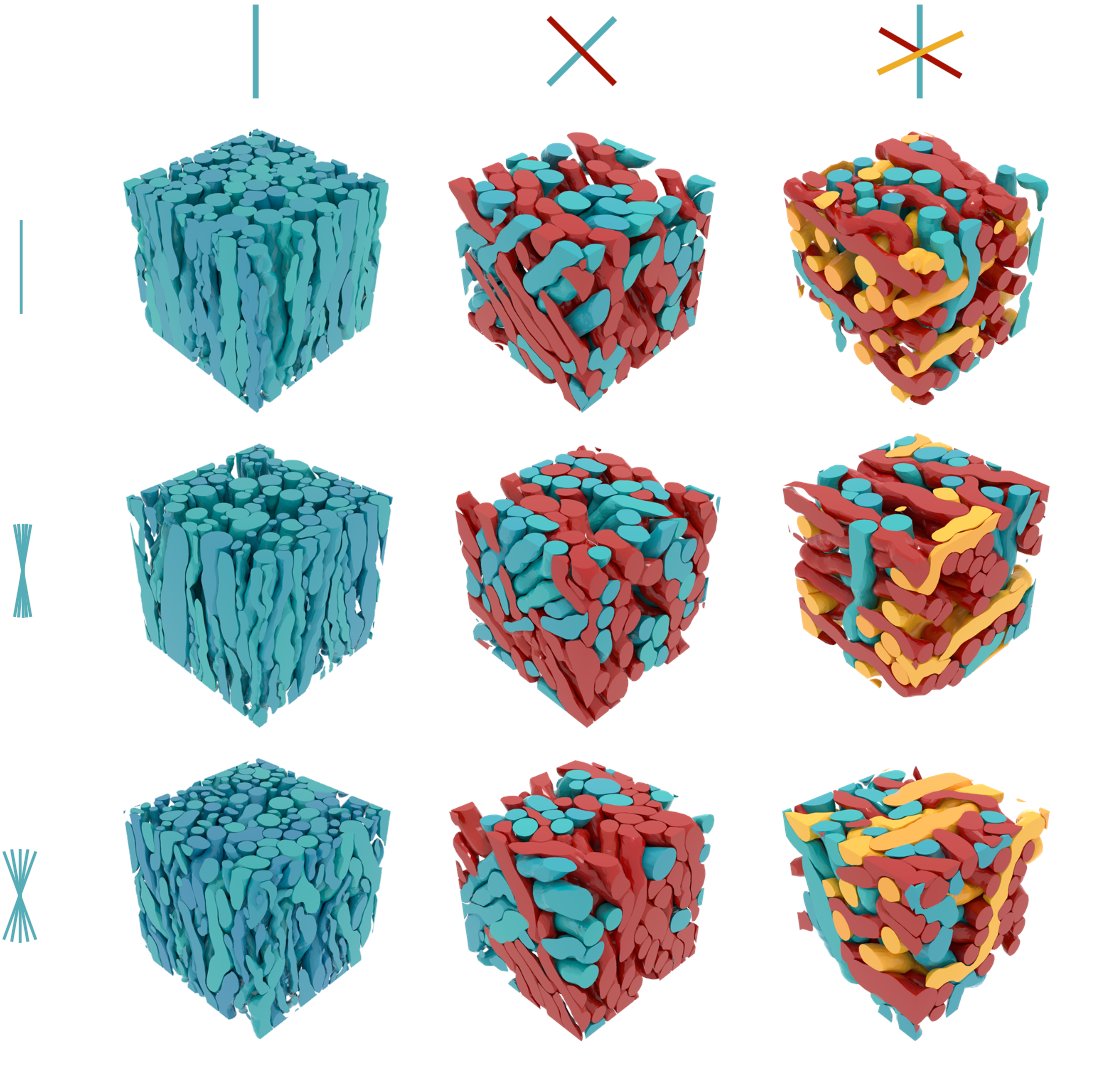
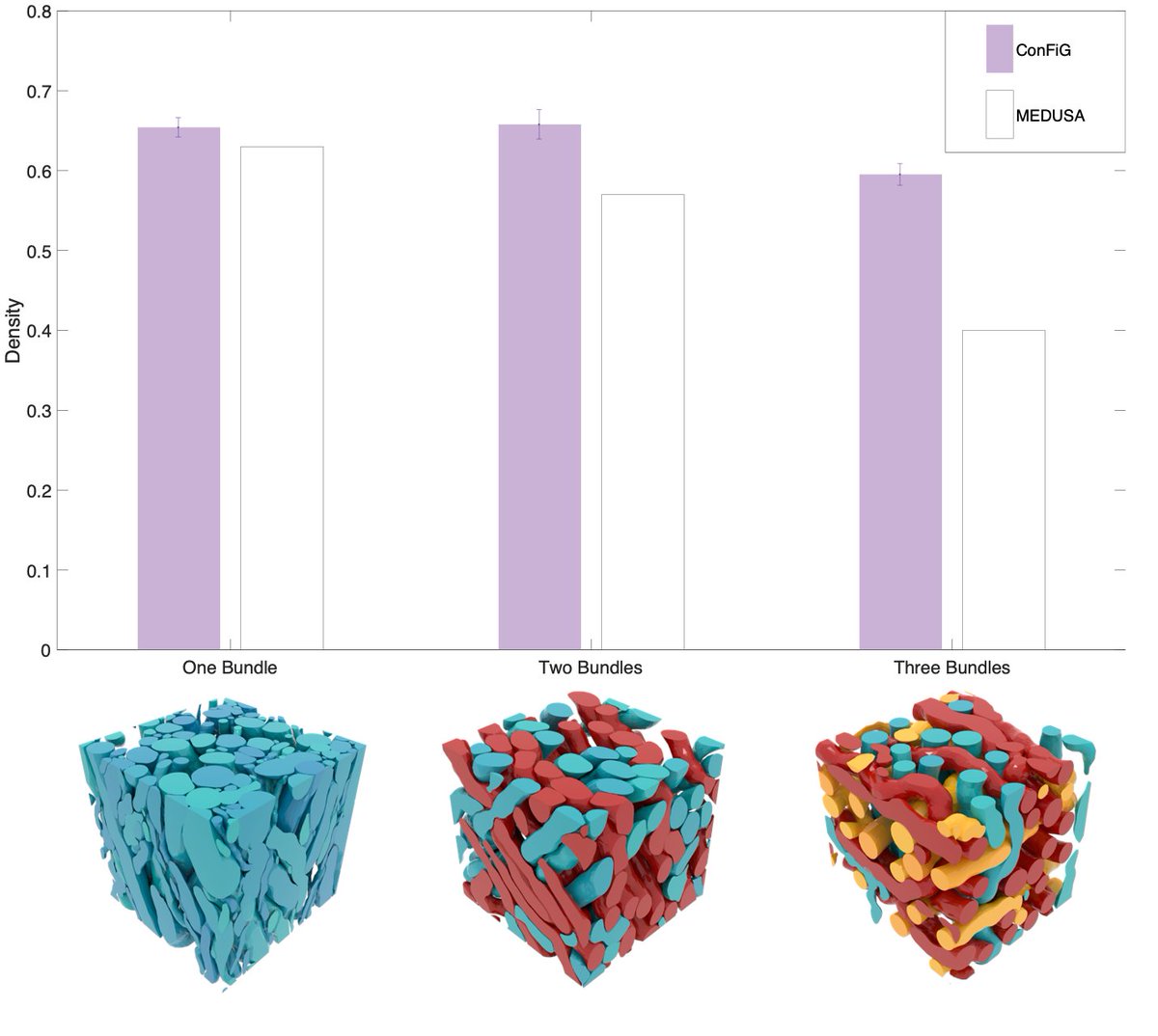
ConFiG can achieve the state-of-the-art packing density in complex WM phantoms including up to three crossing bundles of fibres, outperforming the previous state-of-the-art, MEDUSA (2/8)
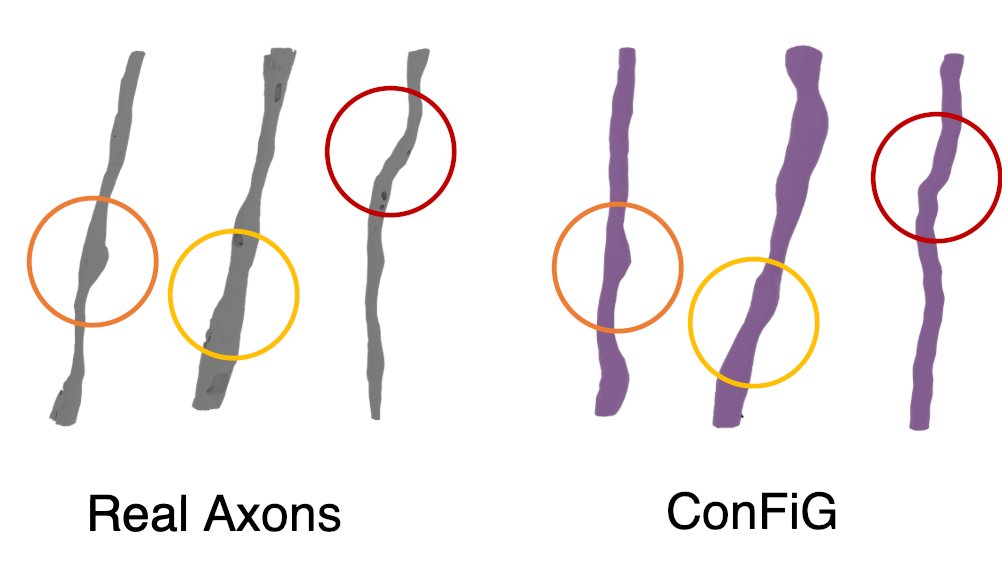
Not only can we generate high density phantoms, but ConFiG phantoms also capture microstructural features found in real axons including fine details like bends and bulges (3/8)
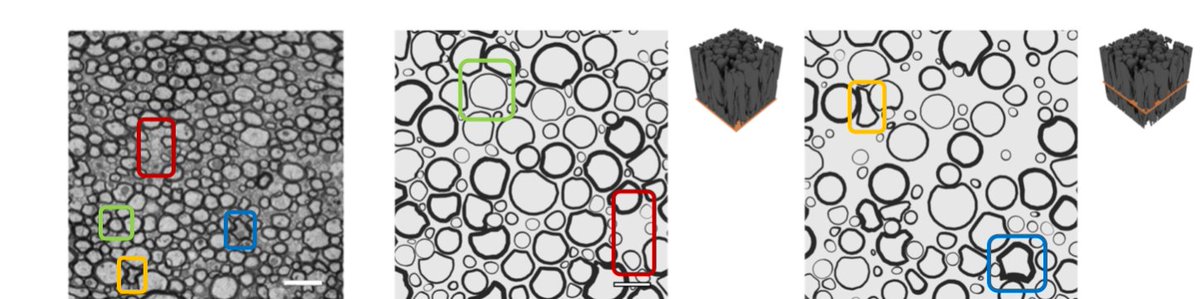
Moreover, ConFiG captures complex fibre cross-sections that we see in real axons, including axons being deformed around one another and naturally occurring pockets of space between axons (4/8)
The realistic microstructure generated by ConFiG is exemplified in this video sweeping though slices of virtual histology from a ConFiG phantom. https://vimeo.com/402472645?ref=tw-share (5/8)
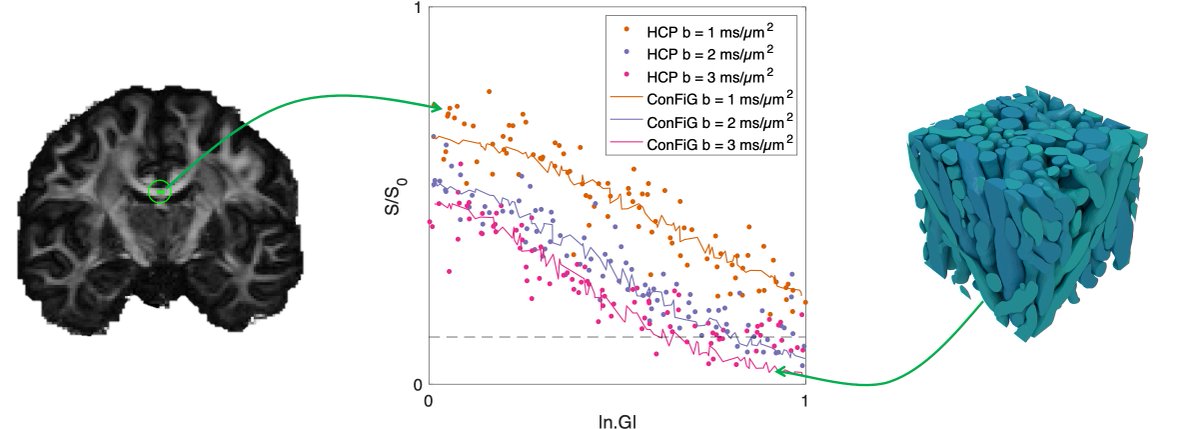
This microstructural realism carries over to the simulated diffusion MRI signal. We show that the simulated signal from ConFiG phantoms made to represent WM regions matches real signal well (6/8)
You might be wondering how we make this happen – we tried to learn from nature, mimicking the way axons grow in real life. This video gives an overview of the ConFiG growth algorithm https://vimeo.com/433661018?ref=tw-share (7/8)
We hope to make ConFiG a useful tool for the wider community to validate and develop new microstructure imaging techniques. We’re working on tidying up the code for release – keep an eye on https://rcallagh.github.io for updates (8/8)

 Read on Twitter
Read on Twitter