I am glad to finally share this huge effort by @nude_mole_rat and the culmination of a great collaboration with @psalmotoxin that started nearly 6 years ago as a cool idea for a side project ..... https://www.nature.com/articles/s41586-020-2410-x
Naked mole-rats (NMR) are known for their resistance to tumour development. Understanding why could help us develop cancer prevention approaches. However, little is known about why they are resistant.
In 2013 two things happened that gave us an idea of how to tackle this question in a systemic way.
First, whole genome CRISPR screens became possible
https://science.sciencemag.org/content/343/6166/84
https://science.sciencemag.org/content/343/6166/80 https://www.nature.com/articles/nbt.2800
First, whole genome CRISPR screens became possible
https://science.sciencemag.org/content/343/6166/84
https://science.sciencemag.org/content/343/6166/80 https://www.nature.com/articles/nbt.2800
Second, Tian X. et al have shown that NMR cells make a high molecular mass hyaluronan and that it mediates the cancer resistance of NMRs https://www.nature.com/articles/nature12234
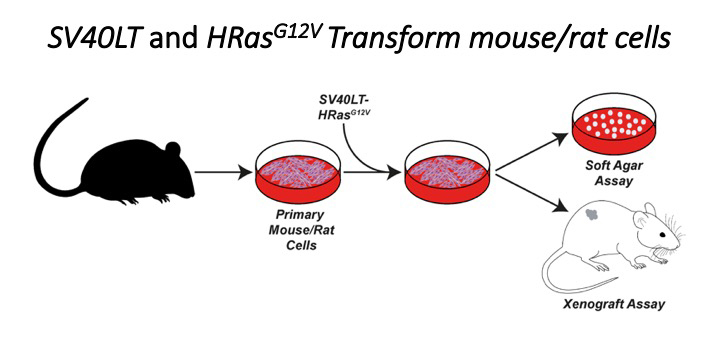
Crucially in this paper the authors used a soft agar assay to show that NMR cells are resistant to transformation by SV40LT and HRAS(G12V) which are enough to transform mouse/rat cells.
They also show that if they perturb hyaluronan production, NMR cells that they can now form colonies in soft agar - i.e. anchorage independent.
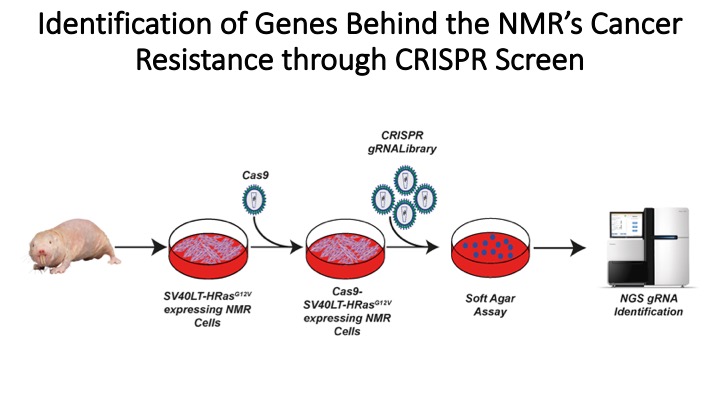
We saw this and thought why not make a NMR whole genome sgRNA library to perform a soft agar screen and identify the genes responsible for tumour resistance is a systemic way.
We made the sgRNA library (not reported here) and also made lentiviral vectors which will allow us to deliver SV40LT, HRAS(G12V) and establish new NMR cell lines and we were ready to go.
Fazal ( @nude_mole_rat) made his first cell lines and used them to practice soft agar assays. To our surprise and contrary to previous data by Tian et al NMR cells expressing SV40LT and HRAS(G12V) formed robust colonies in soft agar.
Our plan was to initially perturb the hyaluronan system as a positive control for our screening strategy. But now we had colonies without any need to disrupt this system: SV40LT and HRAS(G12V) are sufficient to drive tumourigenesis in NMR cells
Our first response was we must have missed something so let's check and repeat this but still got the same results. Fazal was meticulous and he checked the soft agar conditions, temperature, CO2 levels, O2 levels, culturing media....
... he swapped promoters, titrated the lentivirus, linearised the vectors to avoid any viral integration, tried cells from multiple tissues and NMRs. In total he made more than 100 cell lines from 11 NMRs and captured + quantified ~87,000 images!...still the same results.
Importantly, our findings were not restricted to soft agar assays, NMR cells transduced with SV40LT and HRAS(G12V) could be injected into immunodeficient mice and formed turmours.
So what was going on? We used exactly the same protocol as reported by Tian et al to transfect primary NMR and mouse skin cells. However, we saw no anchorage independent growth by NMR or mouse cells.
Tian et al had used multiple transient expression vectors without any antibiotic selection to introduce SV40LT and HRAS(G12V): an extremely inefficient method for introducing oncogenes in an assay lasting 6-weeks!
We are excited to see in their reply to our paper that Tian et al now observe that NMR cells can become turmourigenic through SV40LT and HRAS(G12V) alone!
As for the argument about promotor strength - we used the same promoters as they originally used, as well as linearising plasmids and still observe robust anchorage independent growth.
Conclusions: 1) NMRs are resistant to cancer. 2) NMR cells can be transformed by SV40LT and HRAS(G12V), just like mouse cells. It is important the scientific community are aware of our findings so we can all focus our efforts in understanding why NMRs are remarkable.
Is it the immune system? Mutation rates? Microenvironment? or a combination of all these things. Lots of interesting questions ahead of us to answer!
Well done again to @nude_mole_rat – great collaborative effort with @psalmotoxin and massive thank you to @CR_UK and @Gates_Cambridge for funding this study.
@Cambridge_Uni
@Cambridge_Uni

 Read on Twitter
Read on Twitter