1/6) Delighted to say that our study on genomewide association study of human gut microbiome variation is out today @NatureMicrobiol! Great congrats to #davidhughes @RaesLab @Caketin_Wade @rbacigalupe @mruehlemann
https://www.nature.com/articles/s41564-020-0743-8
https://www.nature.com/articles/s41564-020-0743-8
2/6) We used faecal 16S rRNA gene sequences and host genotype data from the Flemish Gut Flora Project #FGFP, #FoCus and #PopGen cohorts to identify persistent genetic variation associated with multiple gut microbial traits.
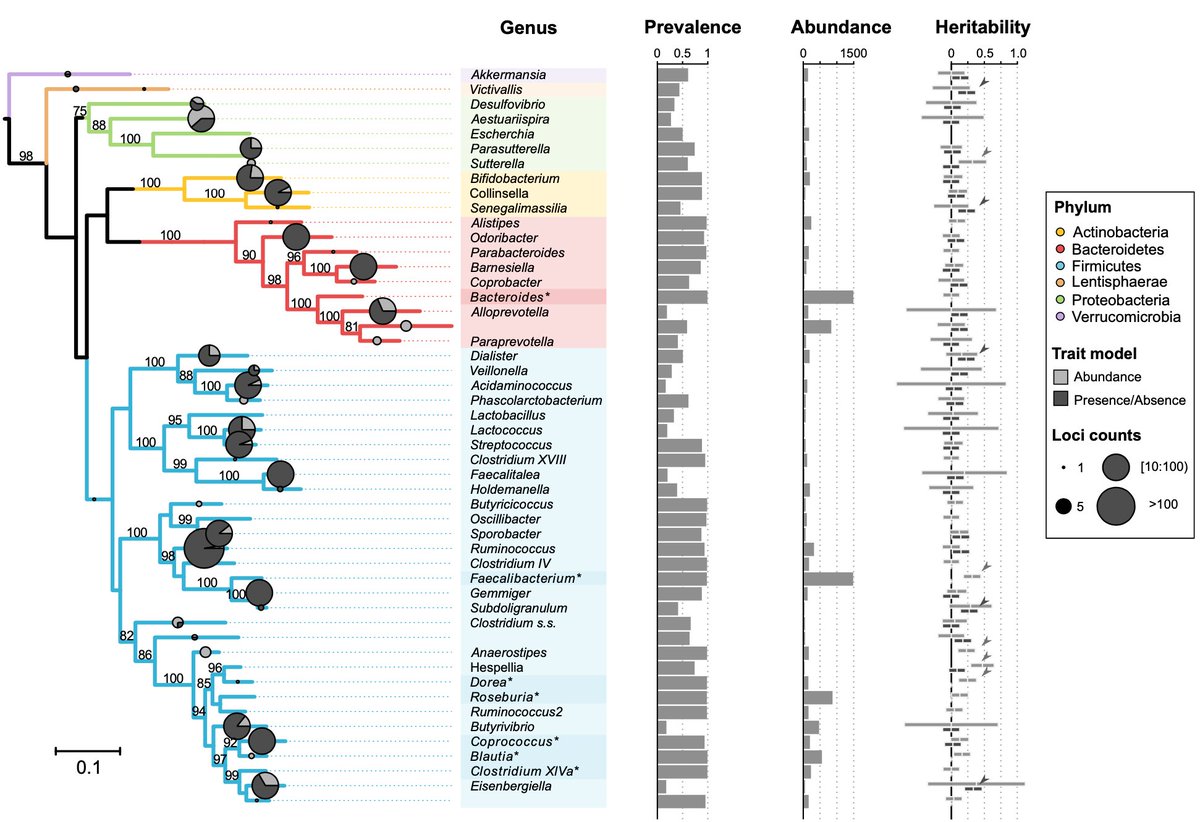
3/6) ...first estimated the proportion of variation in gut microbiota explained by genetic variation (heritability) among individuals in #FGFP. Hespellia, Dorea and Anaerostipes were most heritable bacteria.
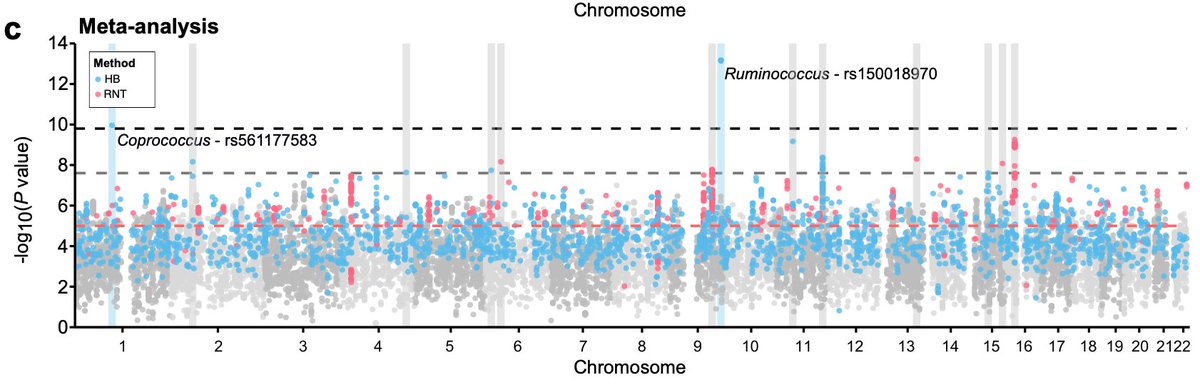
4/6) ...along with our old friend MCM6, identified genetic variants exceeding a study-level p-value (with Ruminococcus and Coprococcus) and a set of variants with persistent and strong evidence of association including the butyrate-producing genus Butyricicoccus.
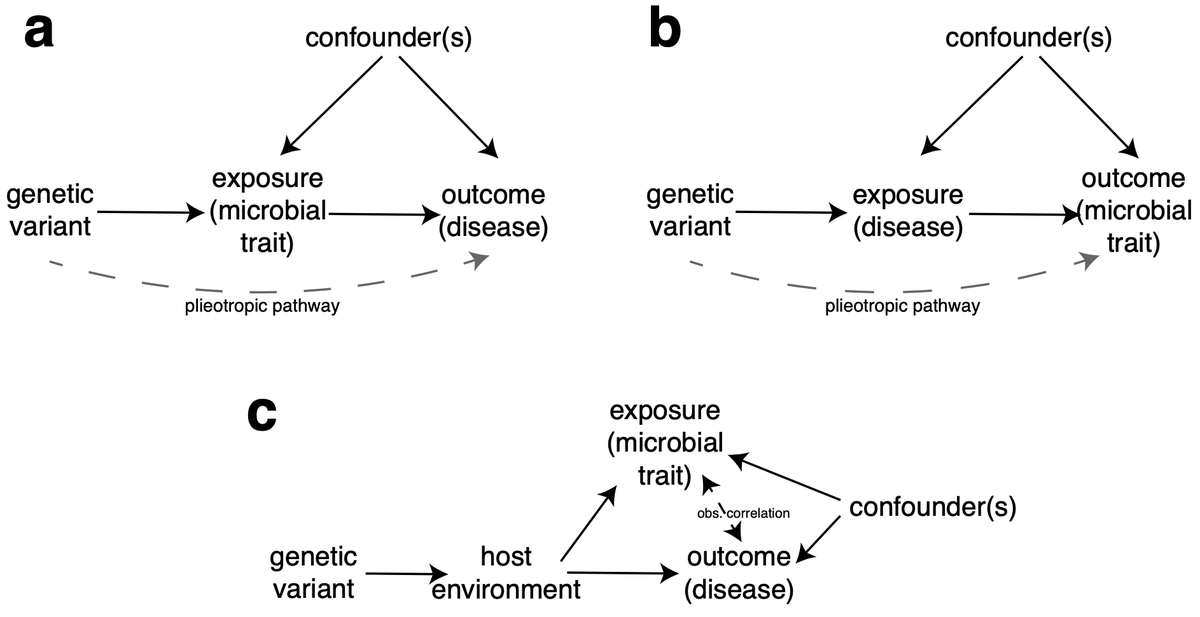
5/6) While associations are compelling the uncertain origin of association (the read out of disease/"environment" or the contributors to it) signals will likely complicate future work looking to dissect function or use associations for causal inference analysis...
(6/6) HUGE thanks to the participants of the studies ( #FGFP, #FoCus and #PopGen) and the team #davidhughes @jeroenraes @Caketin_Wade @rbacigalupe @mruehlemann important work flagging the potential here, but the absolute importance of understanding association before deploying it!

 Read on Twitter
Read on Twitter