Good morning! Today I'll be talking about #MassSpectrometry and how scientists use this method to look examine changes to huge numbers of the #proteins in your #cells to identify how they respond to a #treatment or #infection .
I'll admit my take is somewhat biased - I use this method for looking at #proteins, but it is used to study a range of things such as #drugtesting on athletes, or even on mars landers to look at soil composition. The instrument used in each case is called a mass spectrometer.
So a quick run down on what my day involves today: First we have a meeting for everyone who works on ribosomes in the lab (you'll hear more tomorrow), then I have a single cell experiment to prep (you'll hear this afternoon) and yesterdays experiment to finish up. So lab-heavy!
So first up: whats does a mass spectrometer measure? It looks at 'ions' which are charged molecules. So for me to use a mass spec to look at proteins, I need to take my protein sample and turn it into the ions which can be detected by the mass spec.
The first thing we do is actually to chop up the #proteins we are interested in, with an #enzyme called #trypsin. You can find trypsin in your body in your intestines where it does this same job. Trypsin chops proteins up into small fragments called peptides.
To turn these #peptides into #ions we pass our peptides through a fine glass needle which has a very high voltage applied. This creates a spray which you can see here at the very tip of the needle. In the process, the peptides pick up a charge turning them into #ions.
There’s a tiny hole in the metal cone on the right of the picture above. This is where ions enter the #MassSpec. It’s so small because the inside of the mass spec is kept at high vacuum. Here’s what our mass spec looks like. It’s about 1m tall and weighs 200kg.
The small unit in front of the #MassSpec makes sure the instrument doesn’t see too many ions at once by separating them on a gradient (called liquid chromatography). Think if you won a year of free #pizza, if it all came at once you would throw most away. It’s the same idea.
The manufacturer has made an animation of what happens after ions go into the mass spec: . Theres a lot to take in there so I'll talk through it step by step.
Once the animation gets going (~0:40) the next 50 seconds shows what happens after a set of ions initially enters the #MassPpec. They are focused (0:50) and redirected around the instrument until they enter the C-trap at (1:05). This allows the instrument to collect ions.
At (1:17) they are injected into the orbitrap part of the instrument which is where the different ions are detected. The ions oscillate round the orbitrap, and at (1:40) you can see this oscillation being converted by fourier transform to identify the mass, charge and abundance.
Now from the information we have from the #MassSpec at we have a fair idea of the composition of a peptide in terms of amino acids, but we don't actually know how these amino acids which make up this peptide are ordered - its basically an anagram.
So next, after more ions enter the #MassSpec (1:41), a portion of the instrument called the quadrupole is used to select only a single ion the instrument has decided it is interested in (1:57). It is common to the instrument to select the most abundant few ions at a given time.
The selected ions now go further into the #MassSpec , to a new part called a collision cell. Here they are smashed into fragments (2:07) by colliding with a neutral molecule e.g. nitrogen. By carefully tuning how much you break up your ions, you can create a ladder of fragments.
These are then analysed on the orbitrap before (ends ~2:41)). The #MassSpec is capable of doing some of these steps in parallel, and runs through cycles of the initial scans termed (MS1) and fragmenting and scanning specific ions (called MS2 scans).
We can use the #ion fragments to identify the sequence of the peptide that they came from. The ions extending from the start of the sequence are called b-ions, those extending back from the end of the sequence are called y-ions and are numbered from smallest to largest.
After identifying the peptide sequence, we can then use our knowledge of the proteins made in human cells (or whichever species the sample came from) to reconstruct the proteins that were present in our original sample using computer software design for this.
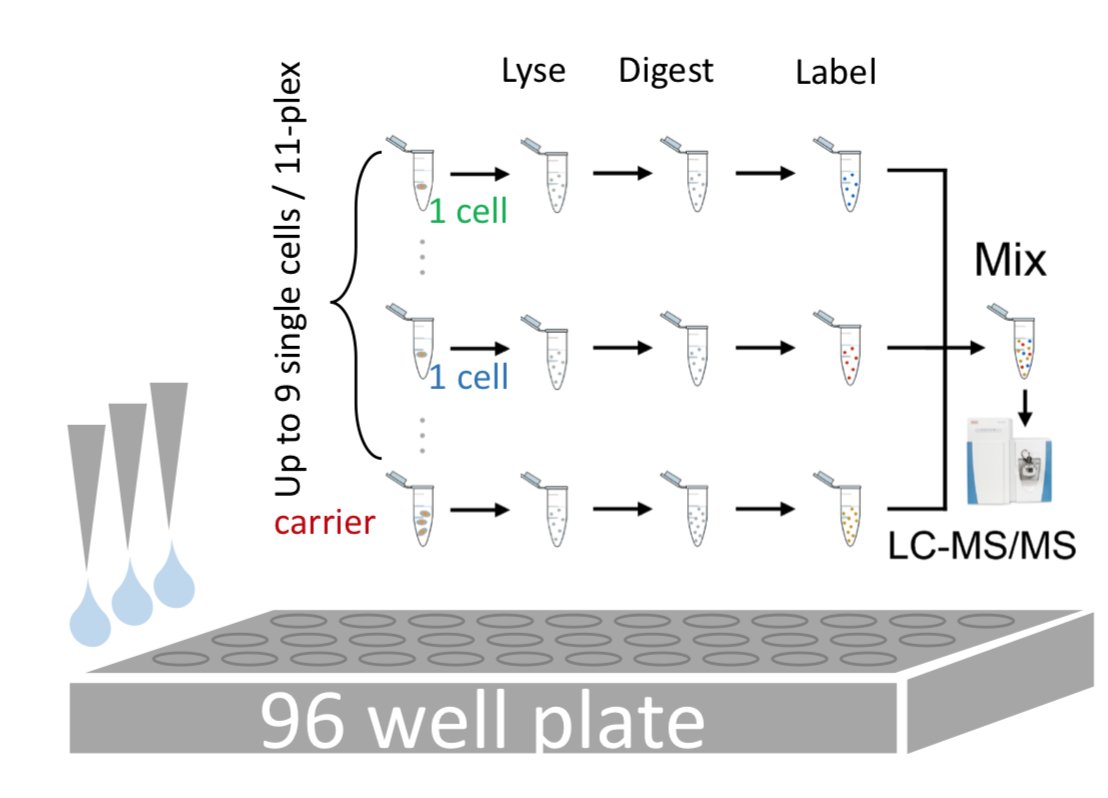
The last thing I’ll be talking about with you today is studying single cells with mass spectrometry, which is what a lot of the work in our lab is focused on. Here are 96 single cells, awaiting their trypsiny fate.
Most studies in proteomics, (and wider cell biology) are done with huge numbers (often over a million) of cells. While these have been a powerful tool, by averaging over such large populations, you can miss important features which are only clear at the individual level.
One of the best methods we have had in the last few years to study this is single cell RNA sequencing. Its used by thousands of labs around the world, and whilst not easy, the methods work well and have been used to study how cells mature or change from one type to another.
There are some downsides to looking at RNA though. Theres often very little of it so our measurements can be noisy, and we are generally interested in the protein that is made from the RNA rather than the RNA itself. The level of the RNA is not always good for estimating this.
Looking at proteins in single cells by mass spec is tricky but now possible. It’s hard because there is very little protein there, and it’s easy to lose the little there is when you process it. Our labs method does it by adding a chemical label to protein from individual cells.
We can then combine this with protein from a much large number of cells which we call a 'carrier'. This helps stop us losing material along the way and gives us enough protein to identify it on the mass spec.
As we use related, but different chemical tags for the peptides from each single cell and the carrier, we can look for the relative levels of these tags in our mass spec data to see how much comes from each cell. We can look at 1000s of peptides from individual cells like this!
A grad student in the lab @hspekt recently published an updated version of our method - this is currently a preprint on @biorxiv . I'll be talking about what preprints are on Saturday! For those interested, you can read it free at: https://www.biorxiv.org/content/early/2018/08/25/399774
I hope you've enjoyed todays tweets on #MassSpec, I'll be back tomorrow to tweet more on proteins. I'll be tweeting on how your cells make proteins, and the many ways this is controlled and potentially taken over during infection.

 Read on Twitter
Read on Twitter